-
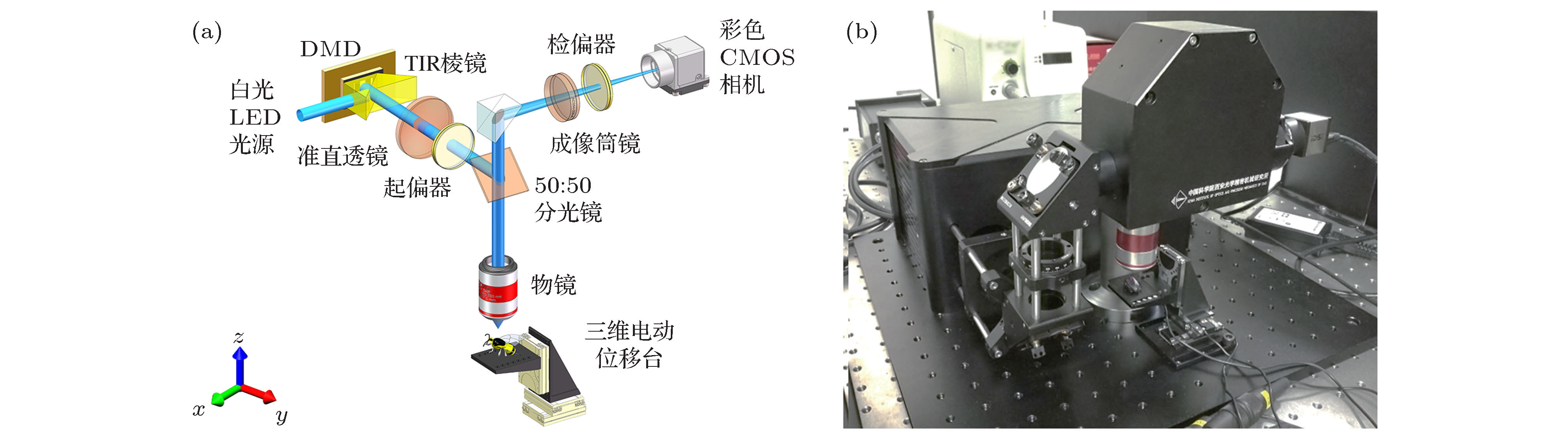
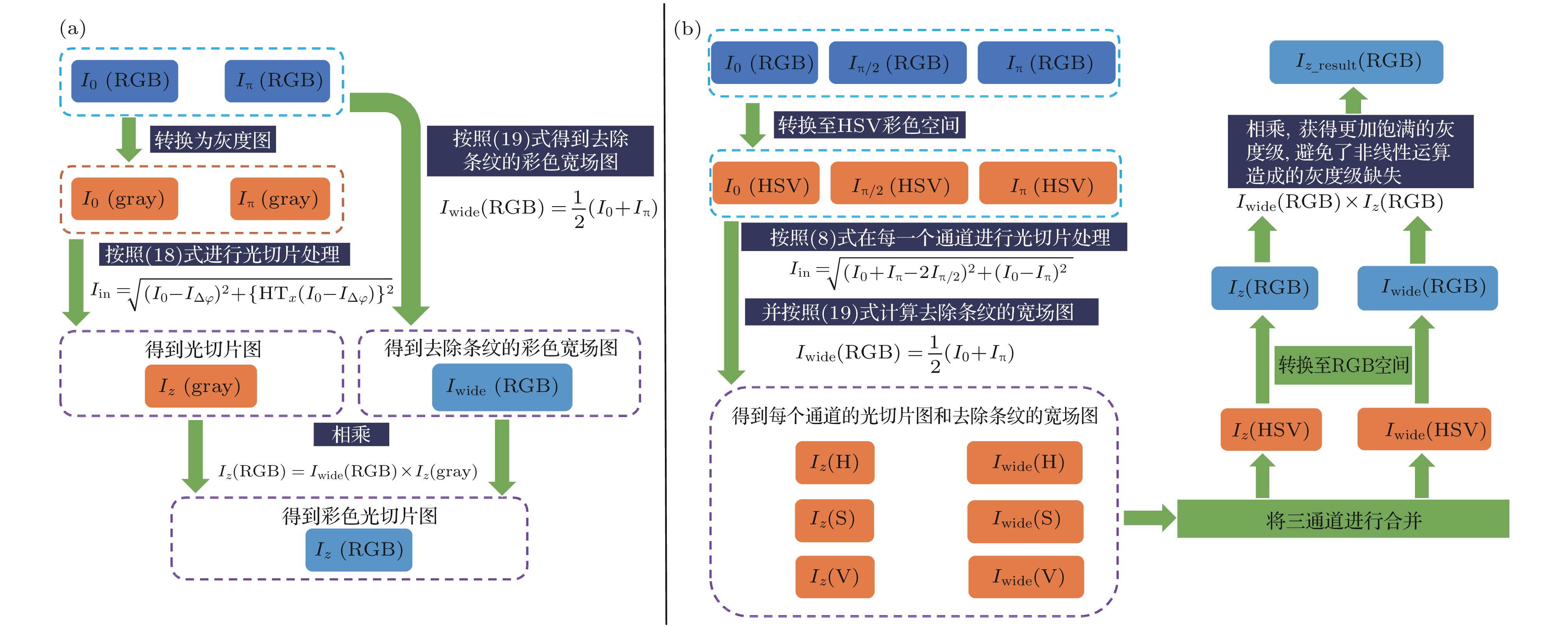
As a wide-field microscopy, structured illumination microscopy (SIM) enables super-resolution and three-dimensional (3D) imaging. It has recently received lots of attention due to the advantages of high spatial resolution, short image recording time, and less photobleaching and phototoxicity. The SIM has found numerous important applications in time-lapse imaging of living tissues and cellular structures in the field of biomedical science. Color information is an important physical quantity describing the characteristics of living creatures and reflects the differences in its microstructure and optical property to some extent. Although HSV (hue, saturation, value) color space based structured illumination full-color 3D optical sectioning technique can recover the full color information on the surface of the samples without color distortion. However, for each optical sectioning, three raw images with fixed phase shift are required to calculate the sectioning images by the rootmean square (RMS) algorithm. This will dramatically increase the data acquisition time and data storage space, especially for a large-scaled sample that needs image stitching strategy. The image processing progress operated in HSV color space need to run the RMS algorithm three times in each channel of HSV space for every section, and transform the images between RGB (red-green-blue) space and HSV space twice. This will absolutely extend the data processing time and put forward higher requirements for computer hardware and software for data storage and processing. To this end, in this paper, a fast 3D color optical sectioning SIM algorithm based on Hilbert-transform is proposed. The Hilbert-transform has proved to be a powerful tool in digital signal and image processing and has successfully applied to the SIM. Here, only two raw images with structured illumination are needed to reconstruct a full-color optical sectioned image for each slice. This fast 3D color sectioning method has the advantage of insensitivity to phase-shift error and has better adaptability to noise, high quality color sectioning images can be obtained under the phase-shift error or noise disturbed environment. The image acquisition data are reduced by 1/3 and the color optical sectioning reconstruction time is saved by about 28%, this new method effectively improves the efficiency and speed for 3D color imaging and will bring a wider application range for SIM.
-
Keywords:
- three-dimensional imaging /
- structured illumination /
- color /
- microscopy
[1] Liu F, Dong B, Liu X, Zheng Y, Zi J 2009 Opt. Express 17 16183
 Google Scholar
Google Scholar
[2] Shevtsova E, Hansson C, Janzen D H, Kjærandsen J 2011 Proc. Natl. Acad. Sci. USA 108 668
 Google Scholar
Google Scholar
[3] Kinoshita S, Yoshioka S 2005 Chem. Phys. Chem. 6 1442
 Google Scholar
Google Scholar
[4] Conchello J A, Lichtman J W 2005 Nat. Methods 2 920
 Google Scholar
Google Scholar
[5] Helmchen F, Denk W 2005 Nat. Methods 2 932
 Google Scholar
Google Scholar
[6] Michels J 2007 J. Microscopy 227 1
 Google Scholar
Google Scholar
[7] Mahou P, Vermot J, Beaurepaire E, Supatto W 2014 Nat. Methods 11 600
 Google Scholar
Google Scholar
[8] Dunn K W, Sandoval R M, Kelly K J, et al. 2002 Am. J. Physiol.-Cell Physiol. 28 C905
 Google Scholar
Google Scholar
[9] Gustafsson M G 2000 J. Microscopy 198 82
 Google Scholar
Google Scholar
[10] Shao L, Kner P, Rego E H, Gustafsson M G 2011 Nat. Methods 8 1044
 Google Scholar
Google Scholar
[11] Kner P, Chhun B B, Griffis E R, Winoto L, Gustafsson M G 2009 Nat. Methods 6 339
 Google Scholar
Google Scholar
[12] Neil M A, Juškaitis R, Wilson T 1997 Opt. Lett. 22 1905
 Google Scholar
Google Scholar
[13] Thomas B, Momany M, Kner P 2013 J. Opt. 15 094004
 Google Scholar
Google Scholar
[14] Mertz J 2011 Nat. Methods 8 811
 Google Scholar
Google Scholar
[15] Dan D, Lei M, Yao B, et al. 2013 Sci. Rep. 3 1116
 Google Scholar
Google Scholar
[16] Karadaglić D, Wilson T 2008 Micron 39 808
 Google Scholar
Google Scholar
[17] Patorski K, Trusiak M, Tkaczyk T 2014 Opt. Express 22 9517
 Google Scholar
Google Scholar
[18] Dan D, Yao B, Lei M 2014 Chin. Sci. Bull. 59 1291
 Google Scholar
Google Scholar
[19] Zhou X, Lei M, Dan D, Yao B, Qian J, Yan S, Yang Y, Min J, Peng T, Ye T 2015 PloS One 10
 Google Scholar
Google Scholar
[20] Qian J, Lei M, Dan D, Yao B, Zhou X, Yang Y, Yan S, Min J, Yu X 2015 Sci. Rep. 5 14513
 Google Scholar
Google Scholar
[21] Qian J, Dang S, Wang Z, Zhou X, Dan D, Yao B, Tong Y, Yang H, Lu Y, Chen Y, Yang X, Bai M, Lei M 2019 Opt. Express 27 4845
 Google Scholar
Google Scholar
[22] 周兴 2018 博士学位论文 (西安: 西安交通大学)
Zhou X 2018 Ph. D. Dissertation (Xi’an: Xi’an Jiaotong University) (in Chinese)
[23] Nayar S K, Nakagawa Y 1990 Proceedings, IEEE International Conference on Robotics and Automation Cincinnati, USA, May 13–18, 1990 p218
[24] Nayar S K, Nakagawa Y 1994 IEEE Trans. Pattern Anal. Mach. Intell. 16 824
 Google Scholar
Google Scholar
-
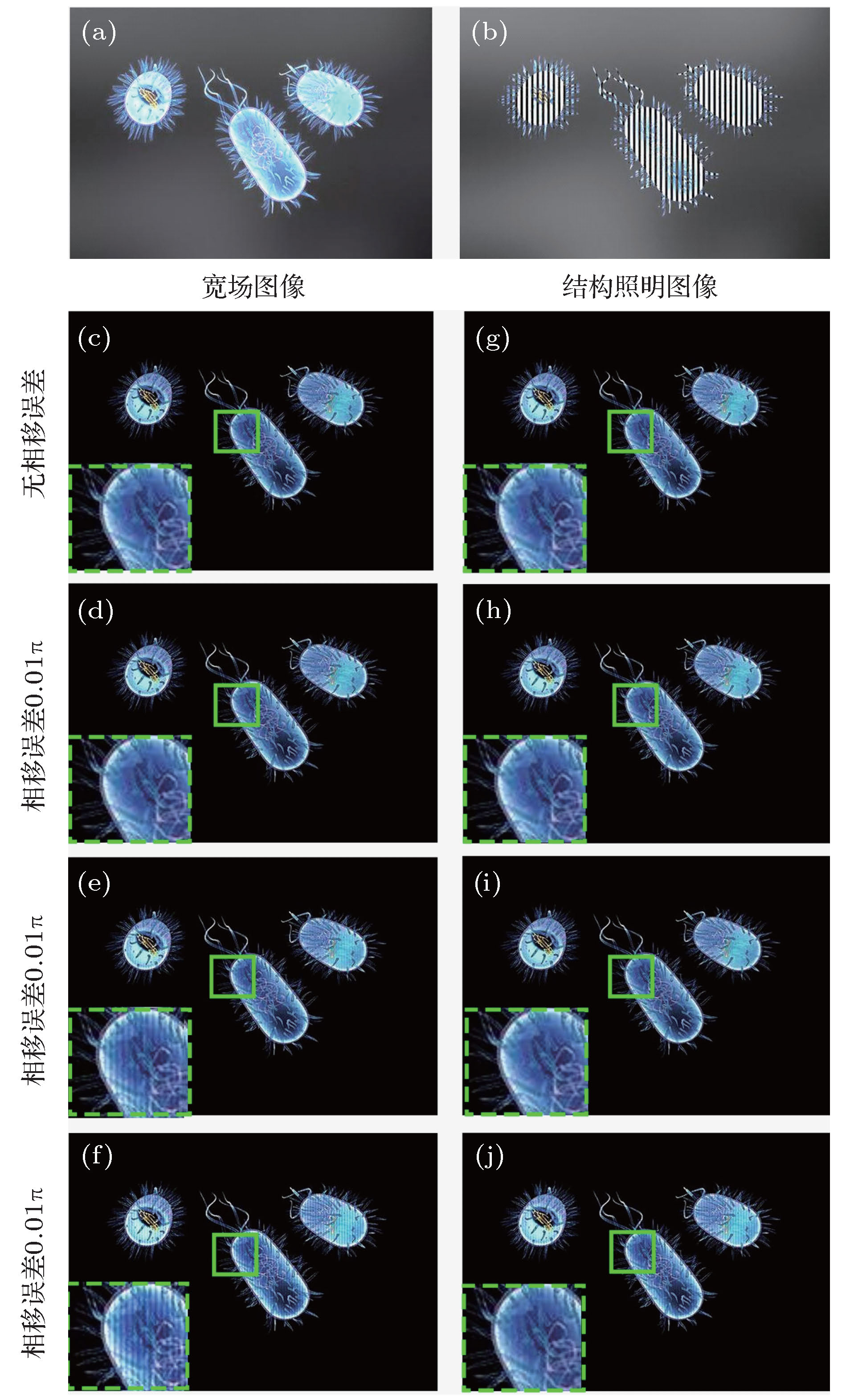
图 2 相移误差对三维光切片图像的影响 (a)含有离焦背景的宽场图像; (b)结构光照明图像; (c)−(f)在不同的相移误差下, HSV-RMS算法所复原的彩色光切片图像; (g)−(j) 不同相移误差下, HT-COS算法所复原的彩色光切片图像
Figure 2. Effect of phase-shift error on optical sectioning images: (a) Wide-field image with defocused background; (b) structured illumination image; (c)−(f) three-dimensional (3D) color optical sectioning images processed by HSV-RMS algorithm under different phase-shift errors; (g)−(j) 3D color optical sectioning images processed by HT-COS algorithm under different phase-shift errors.
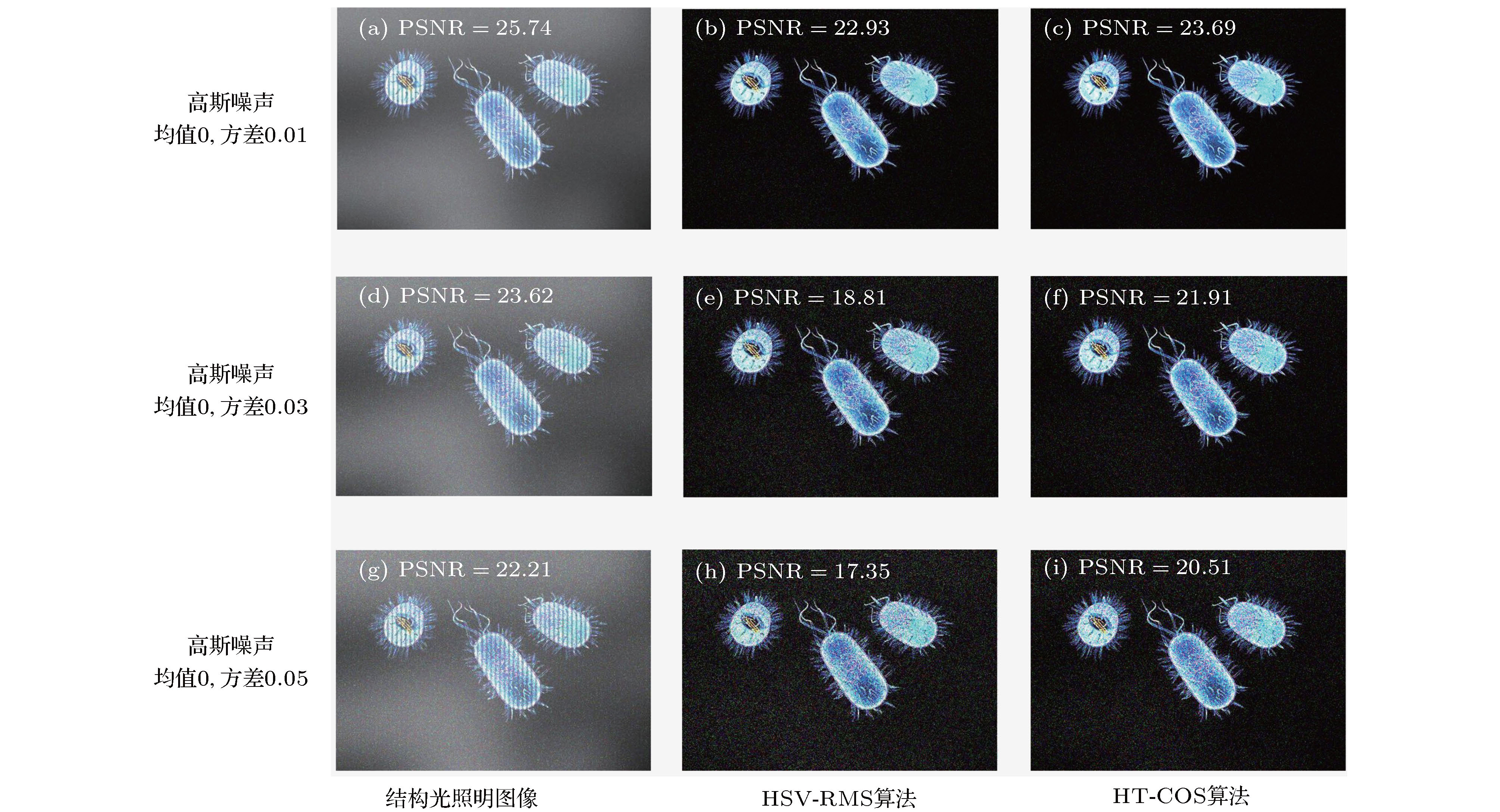
图 3 高斯噪声对光切片重构图像的影响 (a)−(c) 高斯噪声方差为0.01时的结构光照明图像、HSV-RMS算法得到的光切片图像及HT-COS算法得到的光切片图像; (d)−(f) 高斯噪声方差为0.03时的结果; (g)−(i) 高斯噪声方差为0.05时的结果
Figure 3. Influence of Gaussian noise on the reconstructed optical sectioning images. Structured illumination image, optical sectioning images calculated by the HSV-RMS algorithm and HT-COS algorithm, respectively, under the conditions of the Gaussian noise with variances of (a)−(c) 0.01, (d)−(f) 0.03, and (g)−(i) 0.05.
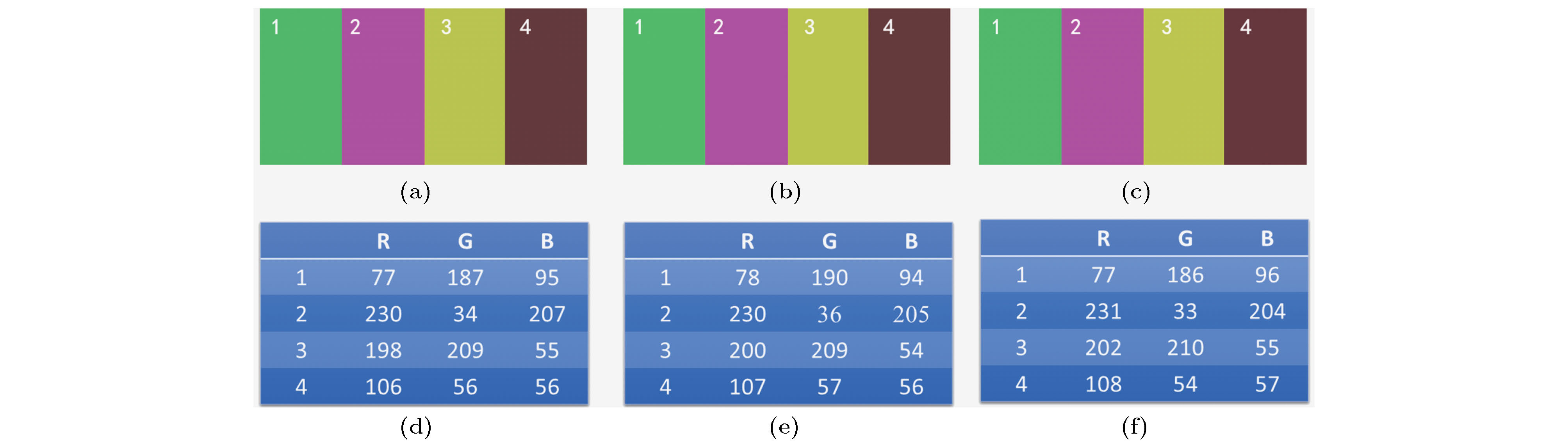
图 4 HSV-RMS算法和HT-COS算法的色彩复原保真度比较 (a)−(c)分别为原始图像、HSV-RMS算法处理后的光切片图像及HT-COS算法处理后的光切片图像; (d)−(f) 3幅图像各色块内的RGB值
Figure 4. Comparison of color restoration fidelity between HSV-RMS algorithm and HT-COS algorithm: (a)−(c) Raw image, optical sectioning image calculated by HSV-RMS algorithm and optical sectioning image calculated by HT-COS algorithm, respectively; (d)−(f) RGB values for the four different regions of each image.
图 6 HSV-RMS算法和HT-COS算法重构的花粉彩色三维光切片图像效果对比 (a) HSV-RMS算法重构的三维图像; (b) HT-COS算法重构的三维图像; (c) 图(a)紫色方框区域的放大图像; (d) 图(b)绿色方框区域内的放大图像; (e) 图(c)中蓝色虚线和图(d)中红色实线上的强度分布; 标尺: 30 μm
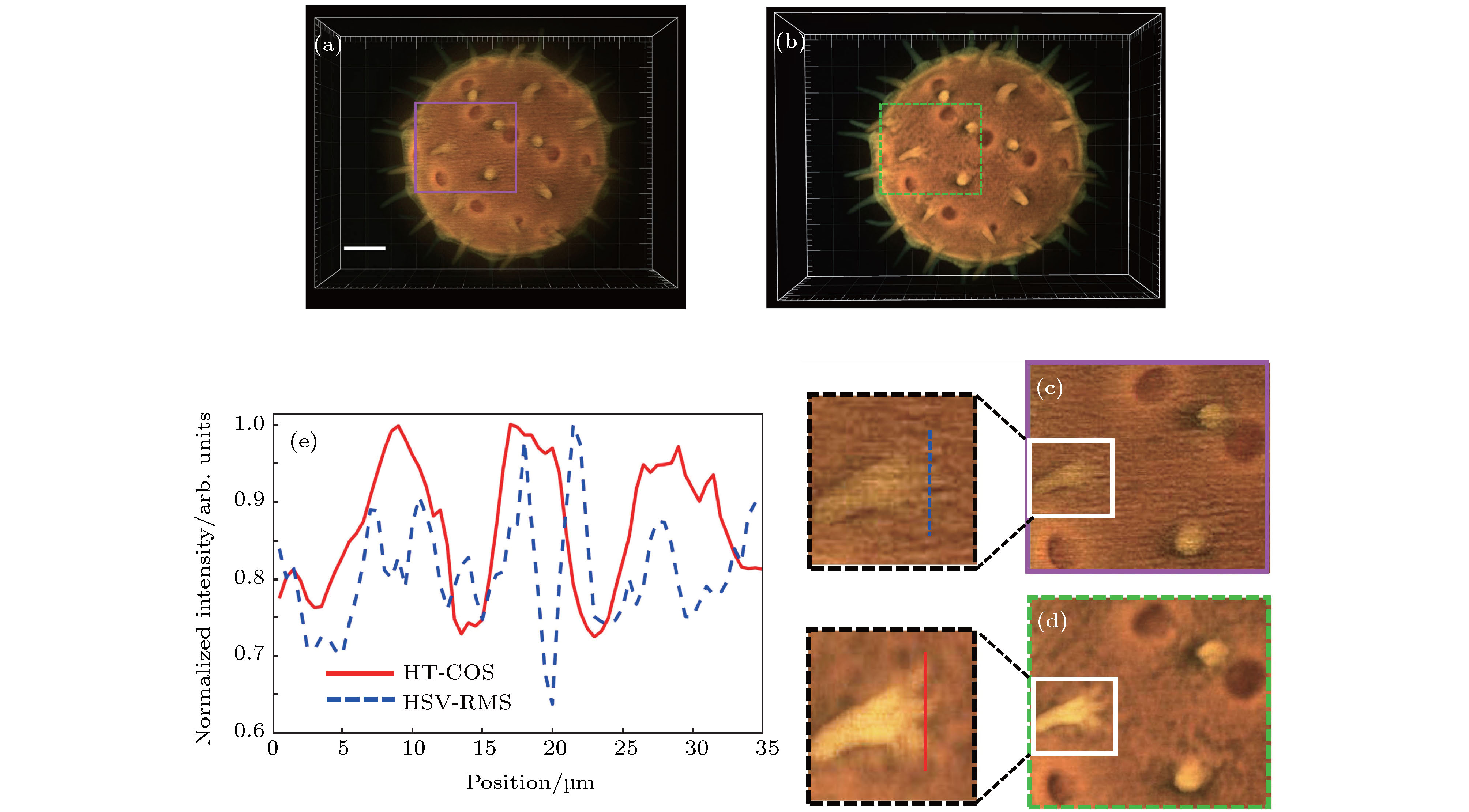
Figure 6. Comparison of reconstructed result of pollen grain between HSV-RMS algorithm and HT-COS algorithm: (a) 3D reconstructed color image from HSV-RMS algorithm; (b) 3D reconstructed color image from HT-COS algorithm; (c) the enlarged image in the purple rectangular box in panel (a); (d) the enlarged image in the green rectangular box in (b); (e) normalized intensity distribution of the line-scan in panel (c) and (d), i.e. the “root-shaped” structure. Scale bar: 30 μm
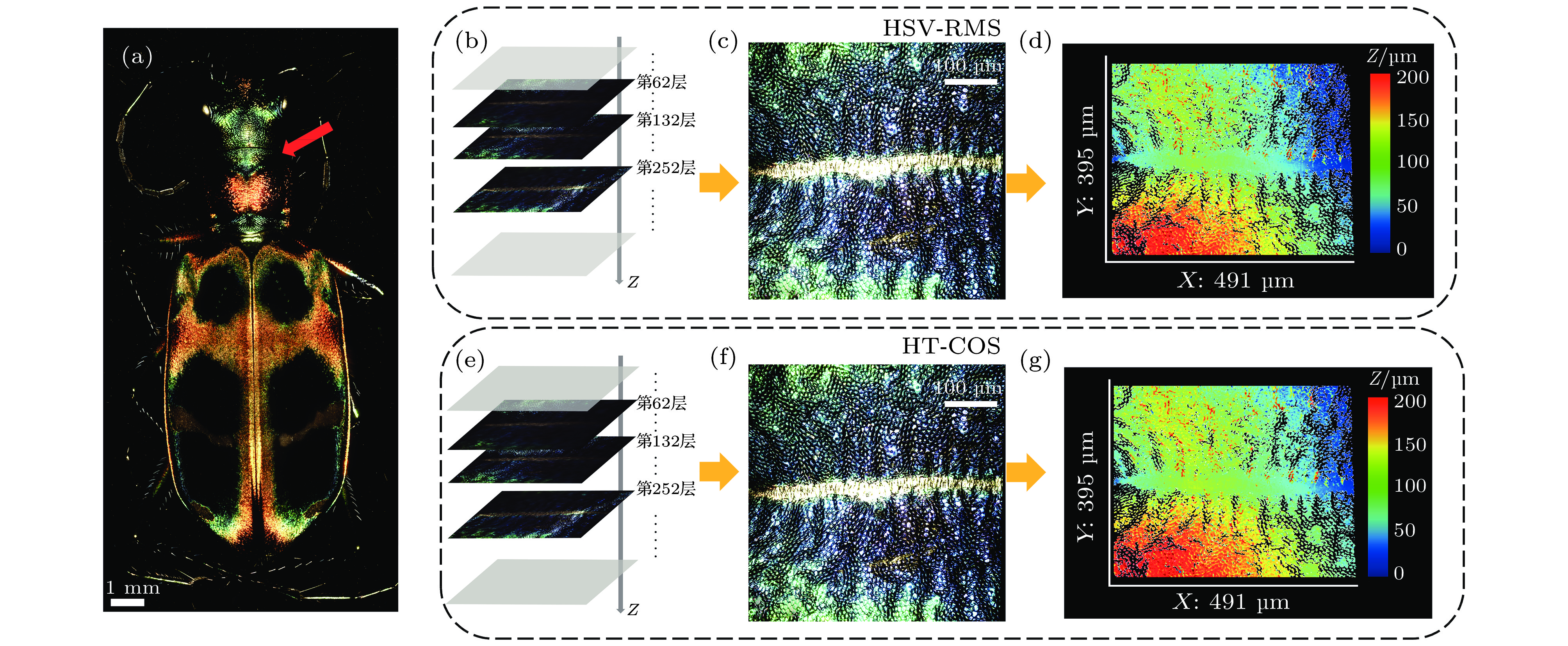
图 7 一种中华虎甲背部的三维彩色SIM成像结果 (a) 该中华虎甲样品完整三维图像的二维最大值投影, 使用4 ×, NA = 0.2物镜拍摄, 共拼接84个视场, 单视场轴向扫描350层; (b) 样品轴向进行三维叠加重构的示意图, 每一层都是经过HSV-RMS算法处理后的光切片图; (c) 图(a)中红色箭头所指区域局部放大的三维光切片最大值投影图像, 使用HSV-RMS算法进行图像处理, 20 ×, NA = 0.45物镜拍摄; (d) 图(c)的三维形貌分布; (e) 样品轴向进行三维叠加重构的示意图, 每一层都是经过HT-COS算法处理后的光切片图; (f)图(a)中红色箭头所指区域局部放大的三维光切片最大值投影图像, 使用HT-COS算法进行图像处理, 20 ×, NA = 0.45物镜拍摄; (g) 图(f)的三维形貌分布
Figure 7. 3D color imaging result of a Chinese tiger beetle: (a) Maximum intensity projection image of the tiger beetle under 4 ×, NA = 0.2 objective lens, the 3D volume is rendered from 84 data sets stitching and sliced 350 layers; (b) schematic diagram of 3D reconstruction in axial direction after imaging processing with HSV-RMS algorithm; (c) maximum intensity projection images of the area pointed by the red arrow in panel (a) processed with HSV-RMS algorithm. The images are captured under 20 ×, NA = 0.45 objective lens; (d) 3D height map of panel (c); (e) schematic diagram of 3D reconstruction in axial direction after imaging processing with HT-COS algorithm; (f) maximum intensity projection images of the area pointed by the red arrow in (a) processed with HT-COS algorithm. The images are captured under 20 ×, NA = 0.45 objective lens; (g) the 3D height map of panel (f).
表 1 两种SIM彩色光切片算法的性能比较
Table 1. Performance comparison of two algorithms for color optical sectioning SIM.
视场个数 图像采集总时间/s 图像处理总时间/s 图像数据容量/GB 三维数据总像素数/pixels HSV-RMS算法 84 1767 13608 1000 1010 HT-COS算法 84 1472 9744 670 1010 -
[1] Liu F, Dong B, Liu X, Zheng Y, Zi J 2009 Opt. Express 17 16183
 Google Scholar
Google Scholar
[2] Shevtsova E, Hansson C, Janzen D H, Kjærandsen J 2011 Proc. Natl. Acad. Sci. USA 108 668
 Google Scholar
Google Scholar
[3] Kinoshita S, Yoshioka S 2005 Chem. Phys. Chem. 6 1442
 Google Scholar
Google Scholar
[4] Conchello J A, Lichtman J W 2005 Nat. Methods 2 920
 Google Scholar
Google Scholar
[5] Helmchen F, Denk W 2005 Nat. Methods 2 932
 Google Scholar
Google Scholar
[6] Michels J 2007 J. Microscopy 227 1
 Google Scholar
Google Scholar
[7] Mahou P, Vermot J, Beaurepaire E, Supatto W 2014 Nat. Methods 11 600
 Google Scholar
Google Scholar
[8] Dunn K W, Sandoval R M, Kelly K J, et al. 2002 Am. J. Physiol.-Cell Physiol. 28 C905
 Google Scholar
Google Scholar
[9] Gustafsson M G 2000 J. Microscopy 198 82
 Google Scholar
Google Scholar
[10] Shao L, Kner P, Rego E H, Gustafsson M G 2011 Nat. Methods 8 1044
 Google Scholar
Google Scholar
[11] Kner P, Chhun B B, Griffis E R, Winoto L, Gustafsson M G 2009 Nat. Methods 6 339
 Google Scholar
Google Scholar
[12] Neil M A, Juškaitis R, Wilson T 1997 Opt. Lett. 22 1905
 Google Scholar
Google Scholar
[13] Thomas B, Momany M, Kner P 2013 J. Opt. 15 094004
 Google Scholar
Google Scholar
[14] Mertz J 2011 Nat. Methods 8 811
 Google Scholar
Google Scholar
[15] Dan D, Lei M, Yao B, et al. 2013 Sci. Rep. 3 1116
 Google Scholar
Google Scholar
[16] Karadaglić D, Wilson T 2008 Micron 39 808
 Google Scholar
Google Scholar
[17] Patorski K, Trusiak M, Tkaczyk T 2014 Opt. Express 22 9517
 Google Scholar
Google Scholar
[18] Dan D, Yao B, Lei M 2014 Chin. Sci. Bull. 59 1291
 Google Scholar
Google Scholar
[19] Zhou X, Lei M, Dan D, Yao B, Qian J, Yan S, Yang Y, Min J, Peng T, Ye T 2015 PloS One 10
 Google Scholar
Google Scholar
[20] Qian J, Lei M, Dan D, Yao B, Zhou X, Yang Y, Yan S, Min J, Yu X 2015 Sci. Rep. 5 14513
 Google Scholar
Google Scholar
[21] Qian J, Dang S, Wang Z, Zhou X, Dan D, Yao B, Tong Y, Yang H, Lu Y, Chen Y, Yang X, Bai M, Lei M 2019 Opt. Express 27 4845
 Google Scholar
Google Scholar
[22] 周兴 2018 博士学位论文 (西安: 西安交通大学)
Zhou X 2018 Ph. D. Dissertation (Xi’an: Xi’an Jiaotong University) (in Chinese)
[23] Nayar S K, Nakagawa Y 1990 Proceedings, IEEE International Conference on Robotics and Automation Cincinnati, USA, May 13–18, 1990 p218
[24] Nayar S K, Nakagawa Y 1994 IEEE Trans. Pattern Anal. Mach. Intell. 16 824
 Google Scholar
Google Scholar
Catalog
Metrics
- Abstract views: 13536
- PDF Downloads: 221
- Cited By: 0














 DownLoad:
DownLoad: