-
p53 is a tumor suppressor protein that plays a crucial role in inhibiting cancer development and maintaining the genetic integrity. Within the cell nucleus, four p53 molecules constitute a stable tetrameric active structure through highly cooperative interactions, bind to DNA via its DNA-binding domain, and transcriptionally activate or inhibit their target genes. However, in most human tumor cells, there are numerous p53 mutations. The majority of these mutations are formed in the p53 DNA-binding domain, importantly, the p53 DNA-binding domain is critical for p53 to form the tetrameric active structures and to regulate the transcription of its downstream target genes. In this work, the all-atom molecular dynamics simulation is conducted to investigate the mechanism of interaction within the wild-type p53 tetramers. This study indicates that the symmetric dimers on either side of the DNA are stable ones, keeping stable structures before and after DNA binding. The binding of two monomers on the same side of the DNA depends on protein-protein interaction provided by two contact surfaces. DNA scaffold stabilizes the tetrameric active structure. Such interactions crucially contribute to the tetramer formation. This study clarifies the internal interactions and key residues within the p53 tetramer in dynamic process, as well as the critical sites at various interaction interfaces. The findings of this study may provide a significant foundation for us to further understand the p53’s anticancer mechanisms, to explore the effective cancer treatment strategies, and in near future, to develop the effective anti-cancer drugs.
-
Keywords:
- p53 core tetramer /
- p53 core domain /
- protein-protein interaction /
- molecular dynamics simulation
[1] Schuijer M, Berns E M 2003 Hum. Mutat. 21 285
 Google Scholar
Google Scholar
[2] Funk W D, Pak D T, Karas R H, Wright W E, Shay J W 1992 Mol. Cell. Biol. 12 2866
[3] Levine A J, Oren M 2009 Nat. Rev. Cancer 9 749
 Google Scholar
Google Scholar
[4] Riley T, Sontag E, Chen P, Levine A 2008 Nat. Rev. Mol. Cell Biol. 9 402
 Google Scholar
Google Scholar
[5] Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian S V, Hainaut P, Olivier M 2007 Hum. Mutat. 28 622
 Google Scholar
Google Scholar
[6] Olivier M, Hollstein M, Hainaut P 2010 CSH Perspect Biol. 2 a001008
 Google Scholar
Google Scholar
[7] Silva J L, Cino E A, Soares I N, Ferreira V F, de Oliveira G A P 2018 Acc. Chem. Res. 51 181
 Google Scholar
Google Scholar
[8] Joerger A, Fersht A R 2007 Oncogene 26 2226
 Google Scholar
Google Scholar
[9] 张丽娟, 晏世伟, 卓益忠 2007 56 2442
 Google Scholar
Google Scholar
Zhang L J, Yan S W, Zhuo Y Z 2007 Acta Phys. Sin. 56 2442
 Google Scholar
Google Scholar
[10] Liu S X, Geng Y Z, Yan S W 2017 Front. Phys. 12 1
 Google Scholar
Google Scholar
[11] 周晗, 耿轶钊, 晏世伟 2023 72 068702
 Google Scholar
Google Scholar
Zhou H, Geng Y Z, Yan S W 2023 Acta Phys. Sin. 72 068702
 Google Scholar
Google Scholar
[12] Gomes A S, Ramos H, Inga A, Sousa E, Saraiva L 2021 Cancers 13 3344
 Google Scholar
Google Scholar
[13] Wang H, Guo M, Wei H, Chen Y 2023 Signal Transduct Target Ther. 8 92
 Google Scholar
Google Scholar
[14] Cho Y, Gorina S, Jeffrey P D, Pavletich N P 1994 Science 265 346
 Google Scholar
Google Scholar
[15] Liu X, Tian W, Cheng J, Li D, Liu T, Zhang L 2020 Comput. Biol. Chem. 84 107194
 Google Scholar
Google Scholar
[16] Tang Y, Yao Y, Wei G 2021 J. Phys. Chem. B 125 10138
[17] Zhao K, Chai X, Johnston K, Clements A, Marmorstein R 2001 J. Biol. Chem. 276 12120
 Google Scholar
Google Scholar
[18] Balagurumoorthy P, Sakamoto H, Lewis M S, Zambrano N, Clore G M, Gronenborn A M, Appella E, Harrington R E 1995 PNAS 92 8591
 Google Scholar
Google Scholar
[19] Nicholls C D, McLure K G, Shields M A, Lee P W 2002 J. Biol. Chem. 277 12937
 Google Scholar
Google Scholar
[20] Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran T E, Shakked Z 2006 Mol. Cell 22 741
 Google Scholar
Google Scholar
[21] McLure K G, Lee P W 1998 EMBO J 17 3342
 Google Scholar
Google Scholar
[22] Weinberg R L, Veprintsev D B, Fersht A R 2004 J. Mol. Biol. 341 1145
 Google Scholar
Google Scholar
[23] Chen Y, Dey R, Chen L 2010 Structure 18 246
 Google Scholar
Google Scholar
[24] Malecka K A, Ho W C, Marmorstein R 2009 Oncogene 28 325
 Google Scholar
Google Scholar
[25] Nagaich A K, Zhurkin V B, Durell S R, Jernigan R L, Appella E, Harrington R E 1999 PNAS 96 1875
 Google Scholar
Google Scholar
[26] Ho W C, Fitzgerald M X, Marmorstein R 2006 J. Biol. Chem. 281 20494
 Google Scholar
Google Scholar
[27] Kamaraj B, Bogaerts A 2015 PLoS One 10 e0134638
 Google Scholar
Google Scholar
[28] Pradhan M R, Siau J W, Kannan S, Nguyen M N, Ouaray Z, Kwoh C K, Lane D P, Ghadessy F, Verma C S 2019 Nucleic Acids Res. 47 1637
 Google Scholar
Google Scholar
[29] Ma B, Pan Y, Gunasekaran K, Venkataraghavan R B, Levine A J, Nussinov R 2005 PNAS 102 3988
 Google Scholar
Google Scholar
[30] Pan Y, Nussinov R 2007 J. Biol. Chem. 282 691
 Google Scholar
Google Scholar
[31] Terakawa T, Takada S 2015 Sci. Rep. 5 17107
 Google Scholar
Google Scholar
[32] Lu Q, Tan Y H, Luo R 2007 J. Phys. Chem. B 111 11538
 Google Scholar
Google Scholar
[33] Abraham M J, Murtola T, Schulz R, Páll S, Smith J C, Hess B, Lindahl E 2015 SoftwareX 1 19
[34] Humphrey W, Dalke A, Schulten K 1996 J. Mol. Graphics 14 33
 Google Scholar
Google Scholar
[35] Arunan E, Desiraju G R, Klein R A, et al. 2011 Pure Appl. Chem. 83 1637
 Google Scholar
Google Scholar
[36] Musafia B, Buchner V, Arad D 1995 J. Mol. Biol. 254 761
 Google Scholar
Google Scholar
[37] Miller III B R, McGee Jr T D, Swails J M, Homeyer N, Gohlke H, Roitberg A E 2012 J. Chem. Theory Comput. 8 3314
 Google Scholar
Google Scholar
[38] Wilcken R, Liu X, Zimmermann M O, Rutherford T J, Fersht A R, Joerger A C, Boeckler F M 2012 J. Am. Chem. Soc. 134 6810
 Google Scholar
Google Scholar
[39] Klein C, Planker E, Diercks T, Kessler H, Kunkele K P, Lang K, Hansen S, Schwaiger M 2001 J. Biol. Chem. 276 49020
 Google Scholar
Google Scholar
[40] Sabapathy K, Lane D P 2018 Nat. Rev. Clin. Oncol. 15 13
 Google Scholar
Google Scholar
[41] Freed-Pastor W A, Prives C 2012 Genes Dev. 26 1268
 Google Scholar
Google Scholar
[42] Dolma L, Muller P A 2022 Cancers 14 5091
 Google Scholar
Google Scholar
[43] Wei H, Qu L, Dai S, et al. 2021 Nat. Commun. 12 2280
 Google Scholar
Google Scholar
[44] Joo W S, Jeffrey P D, Cantor S B, Finnin M S, Livingston D M, Pavletich N P 2002 Genes Dev. 16 583
 Google Scholar
Google Scholar
[45] Gorina S, Pavletich N P 1996 Science 274 1001
 Google Scholar
Google Scholar
[46] Torrie G M, Valleau J P 1977 J. Comput. Phys. 23 187
 Google Scholar
Google Scholar
Torrie G M, Valleau J P 1977 J. Comput. Phys. 23 187
 Google Scholar
Google Scholar
[47] Klein C, Georges G, Kunkele K P, Huber R, Engh R A, Hansen S 2001 J. Biol. Chem. 276 37390
 Google Scholar
Google Scholar
[48] McCammon J A, Harvey S C 1988 Dynamics of Proteins and Nucleic Acids (Cambridge: Cambridge University Press) pp289–302
[49] McCammon J 1984 Rep. Prog. Phys. 47 1
 Google Scholar
Google Scholar
[50] Joerger A C, Fersht A R 2008 Annu. Rev. Biochem. 77 557
 Google Scholar
Google Scholar
-
图 1 p53 核心结合域四聚体结构: A链(蓝色); B链(红色); C链(绿色); D链(橙色). 蓝线表示对称二聚体界面, 红线表示二聚体-二聚体界面
Figure 1. The p53 core binding domain tetramer structure: Chain A (blue); Chain B (red); Chain C (green); Chain D (orange). Blue lines represent symmetric dimer interfaces, while red lines represent dimer-dimer interfaces.
图 2 p53对称二聚体与DNA复合物的初始结构 (a)由A, B链和DNA组成的p53对称二聚体结构; (b) Zn离子的配位结构; (c) DNA轴垂直于图平面的对称二聚体
Figure 2. Initial structure of the p53 symmetric dimer-DNA complex: (a) The p53 symmetric dimer structure composed of chains A, B and DNA; (b) coordination structure of Zn ions; (c) symmetric dimer with the DNA axis perpendicular to the plane of the figure
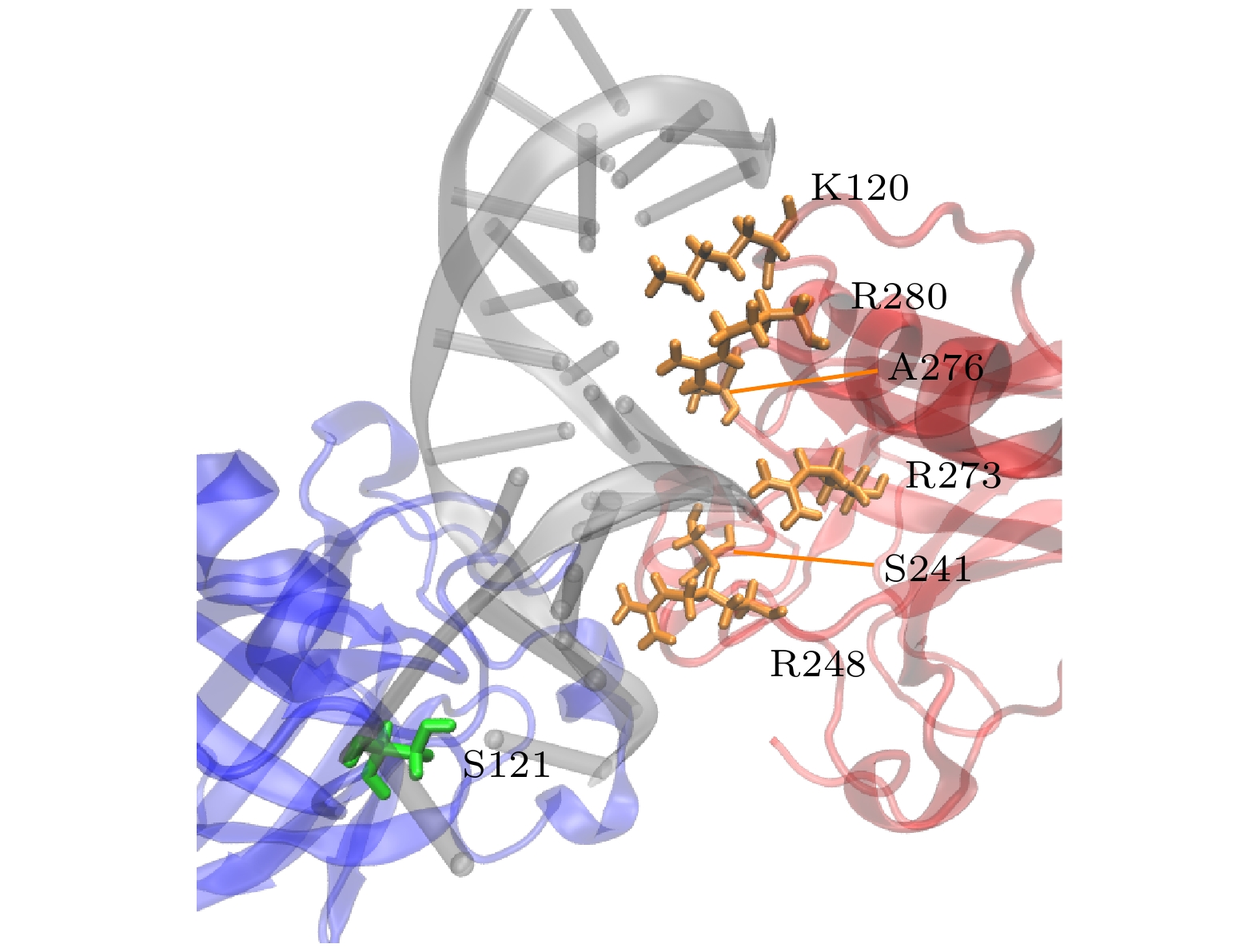
图 7 对称二聚体与DNA结合的关键残基. 黄色残基表示A, B单体与DNA结合的一致残基, 绿色表示与DNA结合不一致的残基
Figure 7. Key residues involved in the binding of the symmetric dimer to DNA. Yellow residues represent consistent residues between monomers A and B and DNA binding, while green residues represent inconsistent residues with DNA binding.
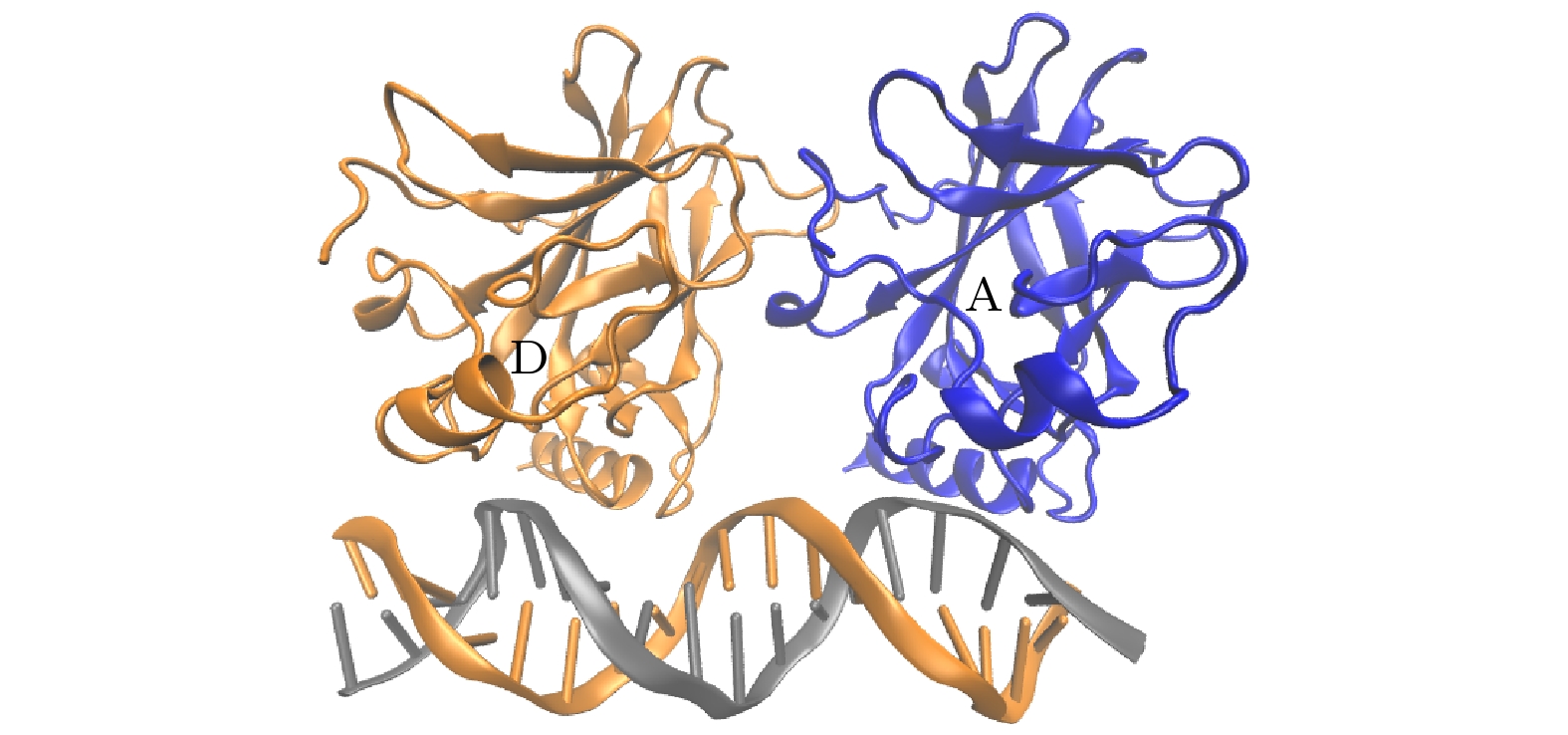
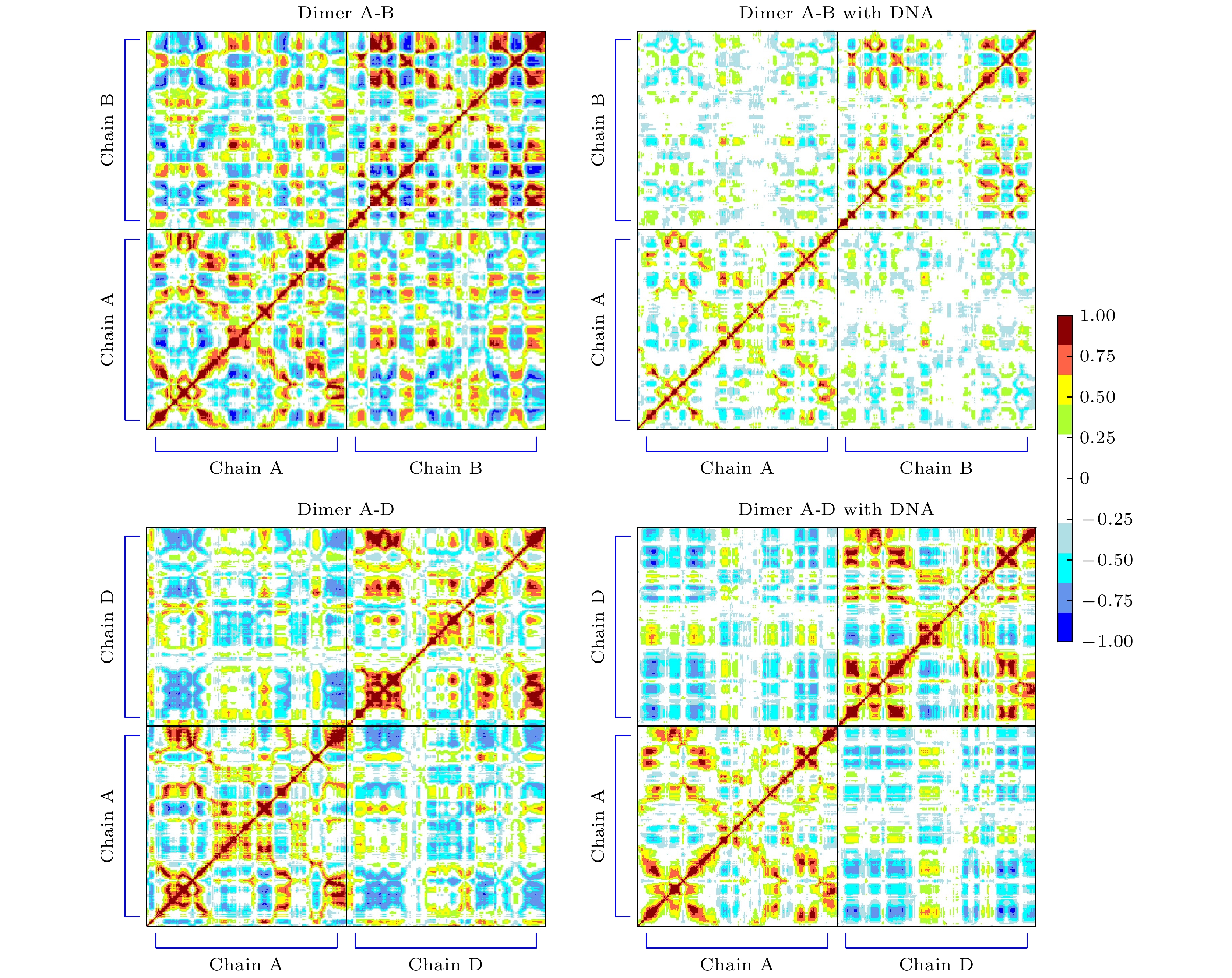
图 9 有/无DNA结合时, A-D二聚体构象上的差别 (a) A, D两个单体质心间距的概率密度分布; (b) A, D分子间接触面积的概率密度分布; (c)有DNA的A, D分子间二聚接触面; (d)无DNA的A, D分子间二聚接触面
Figure 9. Differences in the A-D dimer conformation with/without DNA binding: (a) Probability density distribution of the distance between the centers of mass of A and D monomers; (b) probability density distribution of the interfacial contact area between A and D molecules; (c) dimeric contact area between A and D molecules in the presence of DNA; (d) dimeric contact area between A and D molecules in the absence of DNA.
图 10 (a)无DNA结合时A-D二聚体的RMSD演化; (b) 120 ns前后A-D二聚体的构象重叠, 两单体的构象发生了明显的偏移, 使得A-D之间的相互作用增加. 120 ns前后的构象分别用银色和橙色表示
Figure 10. (a) RMSD evolution of the A-D dimer in the absence of DNA binding; (b) at around 120 ns, there is an overlap in the conformation of the A-D dimer, with noticeable deviations in the conformations of the two monomers, leading to an increased interaction between A and D. Conformations before and after 120 ns are represented in silver and orange, respectively.
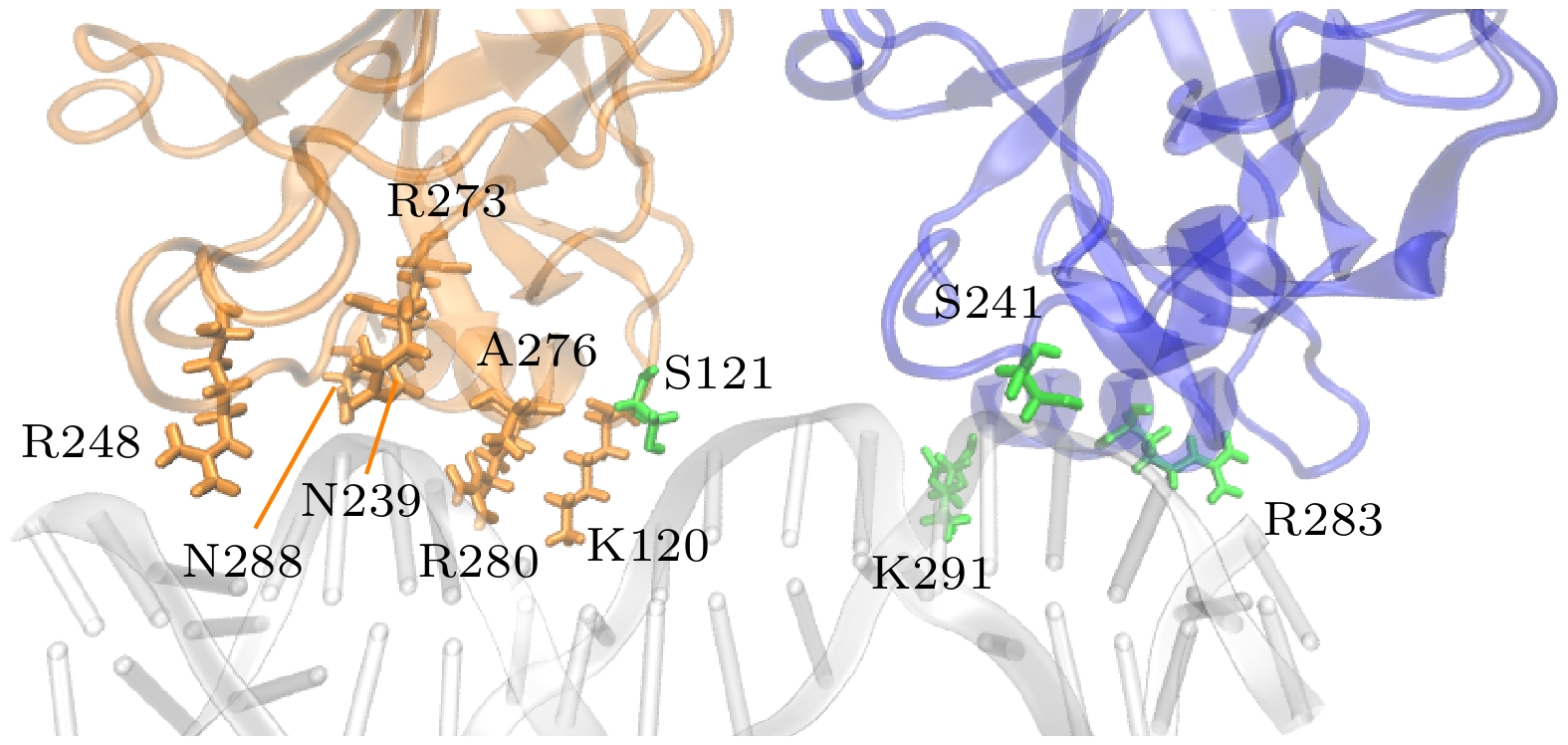
图 15 单体A, D与DNA间相互作用的关键残基. 黄色残基表示A, D单体与DNA结合的一致残基, 绿色表示与DNA结合不一致的残基
Figure 15. Key residues involved in the interaction between monomers A and D with DNA. Yellow residues represent consistent residues between monomers A and D and DNA binding, while green residues represent inconsistent residues with DNA binding.
表 1 氢键、盐桥稳定性
Table 1. Hydrogen bond and salt bridge stability.
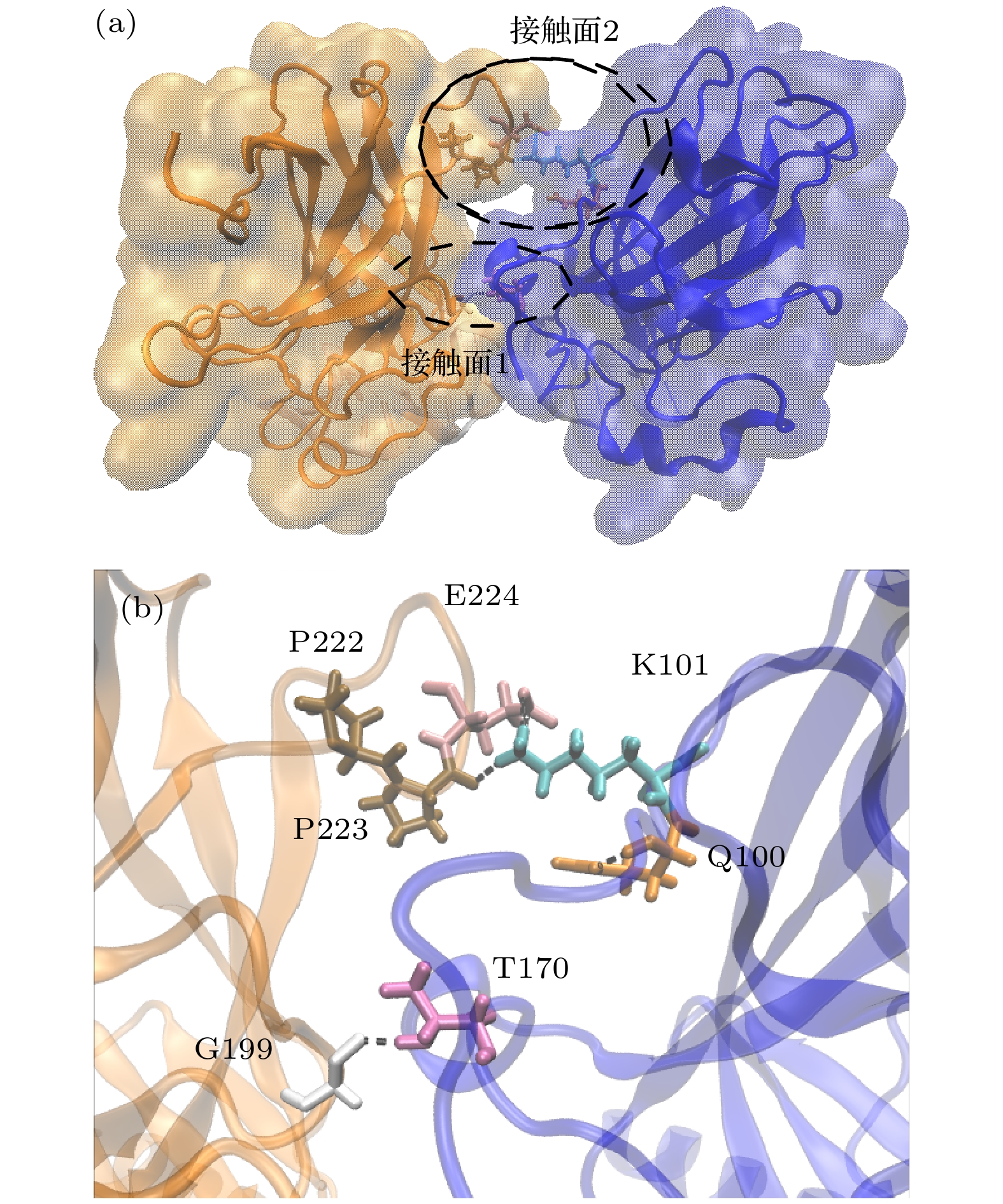
单体A 单体D 占有率/% S94 L201 88.9 N210 D228 55.5 S94 G199 54.7 L264 V225 54.3 S99 D228 35.1, 30.0 S96 T231 33.1 N263 V225 31.8, 26.1 N210 S227 25.1 R267 D228 78.83 (盐桥) R209 E224 35.88 (盐桥) 表 2 与DNA结合时A-D二聚体间的氢键、盐桥稳定性
Table 2. Stability of hydrogen bonds and salt bridges between A-D dimers when binding to DNA.
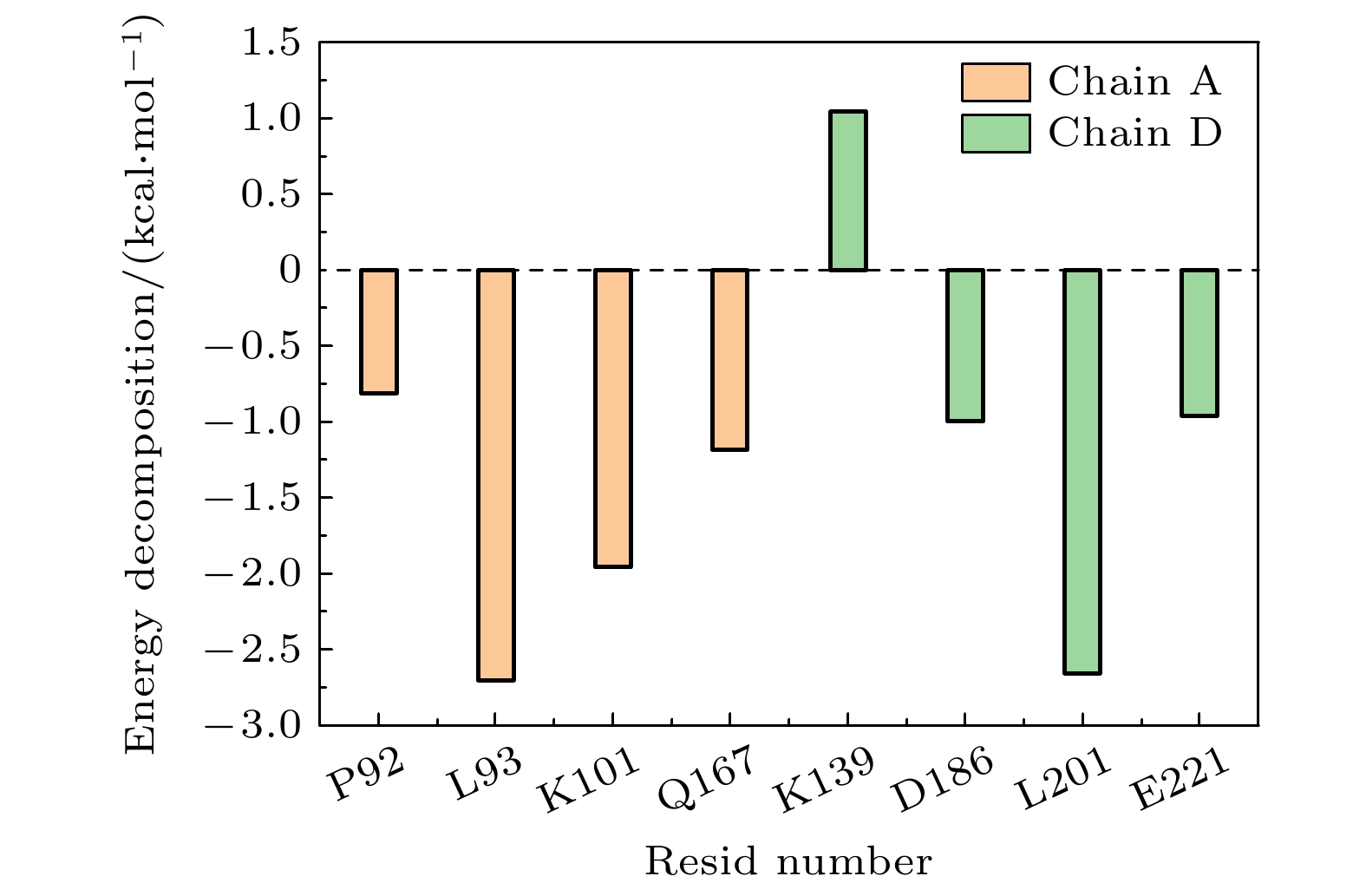
单体A 单体D 持续度/% T170 G199 49.3 K101 P222 19.9 K101 P223 13.0 Q100 P223 16.9 P92 D186 12.6, 12.8 K101 E224 45.89 (盐桥) -
[1] Schuijer M, Berns E M 2003 Hum. Mutat. 21 285
 Google Scholar
Google Scholar
[2] Funk W D, Pak D T, Karas R H, Wright W E, Shay J W 1992 Mol. Cell. Biol. 12 2866
[3] Levine A J, Oren M 2009 Nat. Rev. Cancer 9 749
 Google Scholar
Google Scholar
[4] Riley T, Sontag E, Chen P, Levine A 2008 Nat. Rev. Mol. Cell Biol. 9 402
 Google Scholar
Google Scholar
[5] Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian S V, Hainaut P, Olivier M 2007 Hum. Mutat. 28 622
 Google Scholar
Google Scholar
[6] Olivier M, Hollstein M, Hainaut P 2010 CSH Perspect Biol. 2 a001008
 Google Scholar
Google Scholar
[7] Silva J L, Cino E A, Soares I N, Ferreira V F, de Oliveira G A P 2018 Acc. Chem. Res. 51 181
 Google Scholar
Google Scholar
[8] Joerger A, Fersht A R 2007 Oncogene 26 2226
 Google Scholar
Google Scholar
[9] 张丽娟, 晏世伟, 卓益忠 2007 56 2442
 Google Scholar
Google Scholar
Zhang L J, Yan S W, Zhuo Y Z 2007 Acta Phys. Sin. 56 2442
 Google Scholar
Google Scholar
[10] Liu S X, Geng Y Z, Yan S W 2017 Front. Phys. 12 1
 Google Scholar
Google Scholar
[11] 周晗, 耿轶钊, 晏世伟 2023 72 068702
 Google Scholar
Google Scholar
Zhou H, Geng Y Z, Yan S W 2023 Acta Phys. Sin. 72 068702
 Google Scholar
Google Scholar
[12] Gomes A S, Ramos H, Inga A, Sousa E, Saraiva L 2021 Cancers 13 3344
 Google Scholar
Google Scholar
[13] Wang H, Guo M, Wei H, Chen Y 2023 Signal Transduct Target Ther. 8 92
 Google Scholar
Google Scholar
[14] Cho Y, Gorina S, Jeffrey P D, Pavletich N P 1994 Science 265 346
 Google Scholar
Google Scholar
[15] Liu X, Tian W, Cheng J, Li D, Liu T, Zhang L 2020 Comput. Biol. Chem. 84 107194
 Google Scholar
Google Scholar
[16] Tang Y, Yao Y, Wei G 2021 J. Phys. Chem. B 125 10138
[17] Zhao K, Chai X, Johnston K, Clements A, Marmorstein R 2001 J. Biol. Chem. 276 12120
 Google Scholar
Google Scholar
[18] Balagurumoorthy P, Sakamoto H, Lewis M S, Zambrano N, Clore G M, Gronenborn A M, Appella E, Harrington R E 1995 PNAS 92 8591
 Google Scholar
Google Scholar
[19] Nicholls C D, McLure K G, Shields M A, Lee P W 2002 J. Biol. Chem. 277 12937
 Google Scholar
Google Scholar
[20] Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran T E, Shakked Z 2006 Mol. Cell 22 741
 Google Scholar
Google Scholar
[21] McLure K G, Lee P W 1998 EMBO J 17 3342
 Google Scholar
Google Scholar
[22] Weinberg R L, Veprintsev D B, Fersht A R 2004 J. Mol. Biol. 341 1145
 Google Scholar
Google Scholar
[23] Chen Y, Dey R, Chen L 2010 Structure 18 246
 Google Scholar
Google Scholar
[24] Malecka K A, Ho W C, Marmorstein R 2009 Oncogene 28 325
 Google Scholar
Google Scholar
[25] Nagaich A K, Zhurkin V B, Durell S R, Jernigan R L, Appella E, Harrington R E 1999 PNAS 96 1875
 Google Scholar
Google Scholar
[26] Ho W C, Fitzgerald M X, Marmorstein R 2006 J. Biol. Chem. 281 20494
 Google Scholar
Google Scholar
[27] Kamaraj B, Bogaerts A 2015 PLoS One 10 e0134638
 Google Scholar
Google Scholar
[28] Pradhan M R, Siau J W, Kannan S, Nguyen M N, Ouaray Z, Kwoh C K, Lane D P, Ghadessy F, Verma C S 2019 Nucleic Acids Res. 47 1637
 Google Scholar
Google Scholar
[29] Ma B, Pan Y, Gunasekaran K, Venkataraghavan R B, Levine A J, Nussinov R 2005 PNAS 102 3988
 Google Scholar
Google Scholar
[30] Pan Y, Nussinov R 2007 J. Biol. Chem. 282 691
 Google Scholar
Google Scholar
[31] Terakawa T, Takada S 2015 Sci. Rep. 5 17107
 Google Scholar
Google Scholar
[32] Lu Q, Tan Y H, Luo R 2007 J. Phys. Chem. B 111 11538
 Google Scholar
Google Scholar
[33] Abraham M J, Murtola T, Schulz R, Páll S, Smith J C, Hess B, Lindahl E 2015 SoftwareX 1 19
[34] Humphrey W, Dalke A, Schulten K 1996 J. Mol. Graphics 14 33
 Google Scholar
Google Scholar
[35] Arunan E, Desiraju G R, Klein R A, et al. 2011 Pure Appl. Chem. 83 1637
 Google Scholar
Google Scholar
[36] Musafia B, Buchner V, Arad D 1995 J. Mol. Biol. 254 761
 Google Scholar
Google Scholar
[37] Miller III B R, McGee Jr T D, Swails J M, Homeyer N, Gohlke H, Roitberg A E 2012 J. Chem. Theory Comput. 8 3314
 Google Scholar
Google Scholar
[38] Wilcken R, Liu X, Zimmermann M O, Rutherford T J, Fersht A R, Joerger A C, Boeckler F M 2012 J. Am. Chem. Soc. 134 6810
 Google Scholar
Google Scholar
[39] Klein C, Planker E, Diercks T, Kessler H, Kunkele K P, Lang K, Hansen S, Schwaiger M 2001 J. Biol. Chem. 276 49020
 Google Scholar
Google Scholar
[40] Sabapathy K, Lane D P 2018 Nat. Rev. Clin. Oncol. 15 13
 Google Scholar
Google Scholar
[41] Freed-Pastor W A, Prives C 2012 Genes Dev. 26 1268
 Google Scholar
Google Scholar
[42] Dolma L, Muller P A 2022 Cancers 14 5091
 Google Scholar
Google Scholar
[43] Wei H, Qu L, Dai S, et al. 2021 Nat. Commun. 12 2280
 Google Scholar
Google Scholar
[44] Joo W S, Jeffrey P D, Cantor S B, Finnin M S, Livingston D M, Pavletich N P 2002 Genes Dev. 16 583
 Google Scholar
Google Scholar
[45] Gorina S, Pavletich N P 1996 Science 274 1001
 Google Scholar
Google Scholar
[46] Torrie G M, Valleau J P 1977 J. Comput. Phys. 23 187
 Google Scholar
Google Scholar
Torrie G M, Valleau J P 1977 J. Comput. Phys. 23 187
 Google Scholar
Google Scholar
[47] Klein C, Georges G, Kunkele K P, Huber R, Engh R A, Hansen S 2001 J. Biol. Chem. 276 37390
 Google Scholar
Google Scholar
[48] McCammon J A, Harvey S C 1988 Dynamics of Proteins and Nucleic Acids (Cambridge: Cambridge University Press) pp289–302
[49] McCammon J 1984 Rep. Prog. Phys. 47 1
 Google Scholar
Google Scholar
[50] Joerger A C, Fersht A R 2008 Annu. Rev. Biochem. 77 557
 Google Scholar
Google Scholar
Catalog
Metrics
- Abstract views: 9011
- PDF Downloads: 252
- Cited By: 0















 DownLoad:
DownLoad: