-
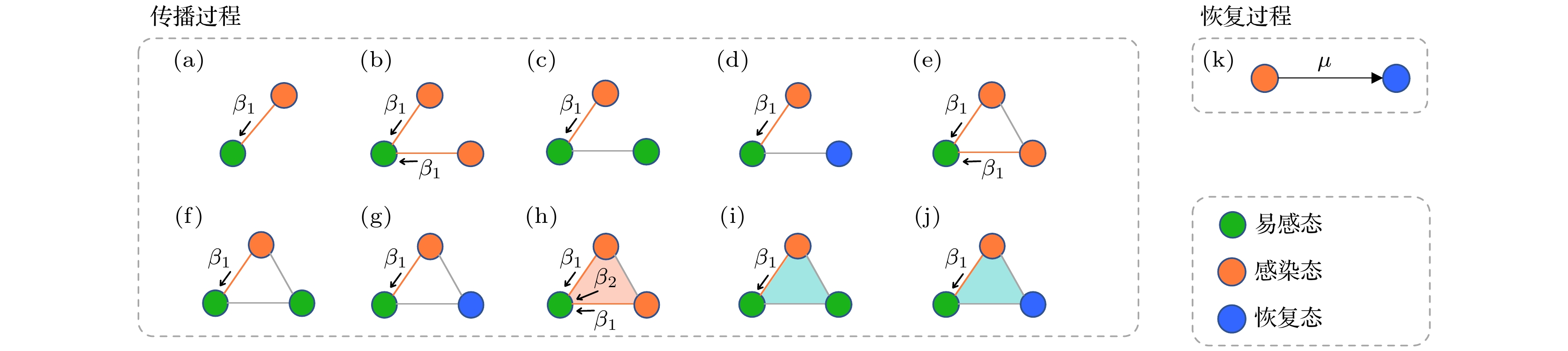
Identifying influential nodes in spreading process in the network is an important step to control the speed and range of spreading, which can be used to accelerate the spread of beneficial information such as healthy behaviors, innovations and suppress the spread of epidemics, rumors and fake news. Existing researches on identification of influential spreaders are mostly based on low-order complex networks with pairwise interactions. However, interactions between individuals occur not only between pairwise nodes but also in groups of three or more nodes, which introduces complex mechanism of reinforcement and indirect influence. The higher-order networks such as simplicial complexes and hypergraphs, can describe features of interactions that go beyond the limitation of pairwise interactions. Currently, there are relatively few researches of identifying influential spreaders in higher-order networks. Some centralities of nodes such as higher-order degree centrality and eigenvector centrality are proposed, but they mostly consider only the network structure. As for identification of influential spreaders, the spreading influence of a node is closely related to the spreading process. In this paper, we work on identification of influential spreaders on simplicial complexes by taking both network structure and dynamical process into consideration. Firstly, we quantitatively describe the dynamics of disease spreading on simplicial complexes by using the Susceptible-Infected-Recovered microscopic Markov equations. Next, we use the microscopic Markov equations to calculate the probability that a node is infected in the spreading process, which is defined as the spreading centrality (SC) of nodes. This spreading centrality involves both the structure of simplicial complex and the dynamical process on it, and is then used to rank the spreading influence of nodes. Simulation results on two types of synthetic simplicial complexes and four real simplicial complexes show that compared with the existing centralities on higher-order networks and the optimal centralities of collective influence and nonbacktracking centrality in complex networks, the proposed spreading centrality can more accurately identify the most influential spreaders in simplicial complexes. In addition, we find that the probability of nodes infected is highly positively correlated with its influence, which is because disease preferentially reaches nodes with many contacts, who can in turn infect their many neighbors and become influential spreaders.
-
Keywords:
- higher-order network /
- simplicial complexes /
- rank the spreading influence of nodes /
- microscopic Markov chain /
- complex networks
[1] Pastor-Satorras R, Vespignani A 2001 Phys. Rev. Lett. 86 3200
 Google Scholar
Google Scholar
[2] Moreno Y, Nekovee M, Pacheco A F 2004 Phys. Rev. E 69 066130
 Google Scholar
Google Scholar
[3] Motter A E 2004 Phys. Rev. Lett. 93 098701
 Google Scholar
Google Scholar
[4] Li D, Fu B, Wang Y, Lu G, Berezin Y, Stanley H E, Havlin S 2014 Proc. Natl. Acad. Sci. 112 669
[5] Kephart J O, Sorkin G B, Chess D M, White S R 1997 Sci. Am. 277 88
[6] Hale T, Angrist N, Goldszmidt R, et al. 2023 Nat. Hum. Behav. 5 529
 Google Scholar
Google Scholar
[7] Rocha Y M, de Moura G A, Desidério G A, et al. 2023 J. Public Health 31 1007
 Google Scholar
Google Scholar
[8] Schäfer B, Witthaut D, Timme M, Latora V 2018 Nat. Commun. 9 1975
 Google Scholar
Google Scholar
[9] 任晓龙, 吕琳媛 2014 科学通报 59 1175
 Google Scholar
Google Scholar
Ren X L, Lü L Y 2014 Sci. Bull. 59 1175
 Google Scholar
Google Scholar
[10] Yang K C, Pierri F, Hui P M, Axelrod D, Torres-Lugo C, Bryden J, Menczer F 2021 Big Data Soc. 8 1
[11] Nielsen B F, Simonsen L, Sneppen K 2021 Phys. Rev. Lett. 126 118301
 Google Scholar
Google Scholar
[12] Freeman L C 1978 Soc. Networks 1 215
 Google Scholar
Google Scholar
[13] Lü L, Zhou T, Zhang Q M, Stanley H E 2016 Nat. Commun. 7 10168
 Google Scholar
Google Scholar
[14] Kitsak M, Gallos L K, Havlin S, Liljeros F, Muchnik L, Stanley H E, Makse H A 2010 Nat. Phys. 6 888
 Google Scholar
Google Scholar
[15] Morone F, Makse H A 2015 Nature 524 65
 Google Scholar
Google Scholar
[16] Sabidussi G 1966 Psychometrika 31 581
 Google Scholar
Google Scholar
[17] Freeman L C 1977 Sociometry 40 35
 Google Scholar
Google Scholar
[18] Estrada E, Rodríguez-Velázquez J A 2005 Phys. Rev. E 71 056103
 Google Scholar
Google Scholar
[19] Bonacich P, Lloyd P 2001 Soc. Networks 23 191
 Google Scholar
Google Scholar
[20] Brin S, Page L 1998 Comput. Netw. ISDN Syst. 30 107
 Google Scholar
Google Scholar
[21] Martin T, Zhang X, Newman M E J 2014 Phys. Rev. E 90 052808
 Google Scholar
Google Scholar
[22] Lü L, Chen D, Ren X L, et al. 2016 Phys. Rep. 650 1
 Google Scholar
Google Scholar
[23] 汪亭亭, 梁宗文, 张若曦 2023 72 048901
 Google Scholar
Google Scholar
Wang T T, Liang Z W, Zhang R X 2023 Acta Phys. Sin. 72 048901
 Google Scholar
Google Scholar
[24] Maji G, Namtirtha A, Dutta A, Malta M C 2020 Exp. Syst. Appl. 144 113092
 Google Scholar
Google Scholar
[25] Liu J Q, Li X R, Dong J C 2021 Sci. China Technol. Sci. 64 451
 Google Scholar
Google Scholar
[26] Liu Y, Zeng Q, Pan L, Tang M 2023 IEEE Trans. Netw. Sci. Eng. 10 2201
 Google Scholar
Google Scholar
[27] Fan T, Lü L, Shi D, Zhou T 2021 Commun. Phys. 4 272
 Google Scholar
Google Scholar
[28] 阮逸润, 老松杨, 汤俊, 白亮, 郭延明 2022 71 176401
 Google Scholar
Google Scholar
Ruan Y R, Lao S Y, Tang J, Bai L, Guo Y M 2022 Acta Phys. Sin. 71 176401
 Google Scholar
Google Scholar
[29] Lung R I, Gaskó N, Suciu M A 2018 Scientometrics 117 1361
 Google Scholar
Google Scholar
[30] Iacopini I, Petri G, Barrat A, Latora V 2019 Nat. Commun. 10 2485
 Google Scholar
Google Scholar
[31] Battiston F, Cencetti G, Iacopini I, Latora V, Lucas M, Patania A, Young J G, Petri G 2020 Phys. Rep. 874 1
 Google Scholar
Google Scholar
[32] de Arruda G F, Petri G, Moreno Y 2020 Phys. Rev. Res. 2 023032
 Google Scholar
Google Scholar
[33] Wang W, Liu Q H, Liang J, Hu Y, Zhou T 2019 Phys. Rep. 820 1
 Google Scholar
Google Scholar
[34] Li W Y, Xue X, Pan L, Lin T, Wang W 2022 Appl. Math. Comput. 412 126595
[35] Fan J, Yin Q, Xia C, Perc M 2022 Proc. R. Soc. A. 478 20220059
 Google Scholar
Google Scholar
[36] Estrada E, Ross G J 2018 J. Theor. Biol. 438 46
 Google Scholar
Google Scholar
[37] Tudisco F, Higham D J 2021 Commun. Phys. 4 201
 Google Scholar
Google Scholar
[38] Kovalenko K, Romance M, Vasilyeva E, et al. 2022 Chaos Solitons Fractals 162 112397
 Google Scholar
Google Scholar
[39] Liu J G, Lin J H, Guo Q, Zhou T 2016 Sci. Rep. 6 21380
 Google Scholar
Google Scholar
[40] Zeng Q, Liu Y, Tang M, Gong J 2021 Knowledge-Based Syst. 229 107365
 Google Scholar
Google Scholar
[41] Li W, Nie Y, Li W, Chen X, Su S, Wang W 2022 Chaos 32 093135
 Google Scholar
Google Scholar
[42] Wang H, Ma C, Chen H S, Lai Y C, Zhang H F 2022 Nat. Commun. 13 3043
 Google Scholar
Google Scholar
[43] Génois M, Barrat A 2018 Epj Data Sci. 7 11
 Google Scholar
Google Scholar
[44] Isella L, Stehlé J, Barrat A, Cattuto C, Pinton J F, Van den Broeck W 2011 J. Theor. Biol. 271 166
 Google Scholar
Google Scholar
[45] Vanhems P, Barrat A, Cattuto C, Pinton J F, Khanafer N, Régis C, Kim B, Comte B, Voirin N 2013 PloS One 8 e73970
 Google Scholar
Google Scholar
[46] Mastrandrea R, Fournet J, Barrat A 2015 PloS One 10 e0136497
 Google Scholar
Google Scholar
-
图 2 节点传播中心性与传播影响力的散点图. $ \overline{{{\mathrm{SC}}}} $与$ \overline{{{\mathrm{S}}}} $分别表示归一化后的节点传播中心性与传播影响力 (a) RSC; (b) SFSC; (c) InVS15; (d) LH10; (e) SFHH; (f) Thiers13
Figure 2. Scatter plots of the spreaing centrality and spreaing influence of nodes. $ \overline{{{\mathrm{SC}}}} $ and $ {\overline{{\mathrm{S}}}} $ represent the normalized spreading centrality and spreading influence of nodes: (a) RSC; (b) SFSC; (c) InVS15; (d) LH10; (e) SFHH; (f) Thiers13.
图 4 不同1阶单纯形传播速率$ \beta_1=\alpha\beta_1^{\mathrm{c}} $下, 节点各中心性与传播影响力的肯德尔相关系数 (a) RSC; (b) SFSC; (c) InVS15; (d) LH10; (e) SFHH; (f) Thiers13
Figure 4. Kendall’s tau correlation of the centralities and the spreading influence of nodes under different 1-simplex spreading rates $ \beta_1=\alpha\beta_1^{\mathrm{c}} $: (a) RSC; (b) SFSC; (c) InVS15; (d) LH10; (e) SFHH; (f) Thiers13
图 5 不同2阶单纯形传播速率$ \beta_2 $下, 节点各中心性与传播影响力的肯德尔相关系数 (a) RSC; (b) SFSC; (c) InVS15; (d) LH10; (e) SFHH; (f) Thiers13
Figure 5. Kendall’s tau correlation of the centralities and the spreading influence of nodes under different 2-simplex spreading rates $ \beta_2 $: (a) RSC; (b) SFSC; (c) InVS15; (d) LH10; (e) SFHH; (f) Thiers13
表 1 合成与真实单纯复形属性
Table 1. Properties of the synthetic and real simplicial complexes
网络 N $\langle k_1\rangle$ $\langle k_2\rangle$ $\beta_1^{\mathrm{c}}$ RSC 2000 20 6 0.045 SFSC 5000 16 5 0.049 InVS15 213 20.19 7.94 0.040 LH10 72 15.94 13.04 0.042 SFHH 403 23.73 8.87 0.026 Thiers13 326 18.10 12.15 0.048 表 2 各中心性的Top-K准确率
Table 2. Top-K accuracy of centralities
网络 $K = 10$ $K = 20$ $K = 30$ $ \mathrm{CI} $ $ \mathrm{NB} $ $ \mathrm{Deg} $ $ \mathrm{EVH} $ $ \mathrm{\mathrm{\mathrm{\mathrm{SC}\mathrm{ }}}} $ $ \mathrm{\mathrm{CI}} $ $ \mathrm{NB} $ $ \mathrm{Deg} $ $ \mathrm{\mathrm{EVH}} $ $ \mathrm{SC} $ $ \mathrm{CI} $ $ \mathrm{NB} $ $ \mathrm{\mathrm{Deg}} $ $ \mathrm{EVH} $ $ \mathrm{SC} $ RSC 0.60 0.60 0.50 0.60 0.60 0.70 0.75 0.60 0.75 0.75 0.70 0.77 0.57 0.77 0.73 SFSC 0.90 0.90 1.00 0.90 0.90 0.95 1.00 0.90 0.95 0.95 0.93 0.97 0.87 0.93 0.97 InVS15 0.40 0.90 0.50 0.70 0.90 0.65 0.90 0.70 0.90 0.90 0.77 0.97 0.70 0.83 0.90 LH10 0.10 1.00 0.90 0.90 0.90 0.55 0.95 0.90 0.95 0.95 0.87 1.00 0.93 0.97 1.00 SFHH 0.80 0.90 0.60 0.70 0.90 0.85 0.90 0.70 0.90 0.90 0.87 0.93 0.77 0.87 0.90 Thiers13 0.30 0.30 0.50 0.10 0.70 0.40 0.40 0.40 0.40 0.70 0.37 0.50 0.43 0.50 0.83 -
[1] Pastor-Satorras R, Vespignani A 2001 Phys. Rev. Lett. 86 3200
 Google Scholar
Google Scholar
[2] Moreno Y, Nekovee M, Pacheco A F 2004 Phys. Rev. E 69 066130
 Google Scholar
Google Scholar
[3] Motter A E 2004 Phys. Rev. Lett. 93 098701
 Google Scholar
Google Scholar
[4] Li D, Fu B, Wang Y, Lu G, Berezin Y, Stanley H E, Havlin S 2014 Proc. Natl. Acad. Sci. 112 669
[5] Kephart J O, Sorkin G B, Chess D M, White S R 1997 Sci. Am. 277 88
[6] Hale T, Angrist N, Goldszmidt R, et al. 2023 Nat. Hum. Behav. 5 529
 Google Scholar
Google Scholar
[7] Rocha Y M, de Moura G A, Desidério G A, et al. 2023 J. Public Health 31 1007
 Google Scholar
Google Scholar
[8] Schäfer B, Witthaut D, Timme M, Latora V 2018 Nat. Commun. 9 1975
 Google Scholar
Google Scholar
[9] 任晓龙, 吕琳媛 2014 科学通报 59 1175
 Google Scholar
Google Scholar
Ren X L, Lü L Y 2014 Sci. Bull. 59 1175
 Google Scholar
Google Scholar
[10] Yang K C, Pierri F, Hui P M, Axelrod D, Torres-Lugo C, Bryden J, Menczer F 2021 Big Data Soc. 8 1
[11] Nielsen B F, Simonsen L, Sneppen K 2021 Phys. Rev. Lett. 126 118301
 Google Scholar
Google Scholar
[12] Freeman L C 1978 Soc. Networks 1 215
 Google Scholar
Google Scholar
[13] Lü L, Zhou T, Zhang Q M, Stanley H E 2016 Nat. Commun. 7 10168
 Google Scholar
Google Scholar
[14] Kitsak M, Gallos L K, Havlin S, Liljeros F, Muchnik L, Stanley H E, Makse H A 2010 Nat. Phys. 6 888
 Google Scholar
Google Scholar
[15] Morone F, Makse H A 2015 Nature 524 65
 Google Scholar
Google Scholar
[16] Sabidussi G 1966 Psychometrika 31 581
 Google Scholar
Google Scholar
[17] Freeman L C 1977 Sociometry 40 35
 Google Scholar
Google Scholar
[18] Estrada E, Rodríguez-Velázquez J A 2005 Phys. Rev. E 71 056103
 Google Scholar
Google Scholar
[19] Bonacich P, Lloyd P 2001 Soc. Networks 23 191
 Google Scholar
Google Scholar
[20] Brin S, Page L 1998 Comput. Netw. ISDN Syst. 30 107
 Google Scholar
Google Scholar
[21] Martin T, Zhang X, Newman M E J 2014 Phys. Rev. E 90 052808
 Google Scholar
Google Scholar
[22] Lü L, Chen D, Ren X L, et al. 2016 Phys. Rep. 650 1
 Google Scholar
Google Scholar
[23] 汪亭亭, 梁宗文, 张若曦 2023 72 048901
 Google Scholar
Google Scholar
Wang T T, Liang Z W, Zhang R X 2023 Acta Phys. Sin. 72 048901
 Google Scholar
Google Scholar
[24] Maji G, Namtirtha A, Dutta A, Malta M C 2020 Exp. Syst. Appl. 144 113092
 Google Scholar
Google Scholar
[25] Liu J Q, Li X R, Dong J C 2021 Sci. China Technol. Sci. 64 451
 Google Scholar
Google Scholar
[26] Liu Y, Zeng Q, Pan L, Tang M 2023 IEEE Trans. Netw. Sci. Eng. 10 2201
 Google Scholar
Google Scholar
[27] Fan T, Lü L, Shi D, Zhou T 2021 Commun. Phys. 4 272
 Google Scholar
Google Scholar
[28] 阮逸润, 老松杨, 汤俊, 白亮, 郭延明 2022 71 176401
 Google Scholar
Google Scholar
Ruan Y R, Lao S Y, Tang J, Bai L, Guo Y M 2022 Acta Phys. Sin. 71 176401
 Google Scholar
Google Scholar
[29] Lung R I, Gaskó N, Suciu M A 2018 Scientometrics 117 1361
 Google Scholar
Google Scholar
[30] Iacopini I, Petri G, Barrat A, Latora V 2019 Nat. Commun. 10 2485
 Google Scholar
Google Scholar
[31] Battiston F, Cencetti G, Iacopini I, Latora V, Lucas M, Patania A, Young J G, Petri G 2020 Phys. Rep. 874 1
 Google Scholar
Google Scholar
[32] de Arruda G F, Petri G, Moreno Y 2020 Phys. Rev. Res. 2 023032
 Google Scholar
Google Scholar
[33] Wang W, Liu Q H, Liang J, Hu Y, Zhou T 2019 Phys. Rep. 820 1
 Google Scholar
Google Scholar
[34] Li W Y, Xue X, Pan L, Lin T, Wang W 2022 Appl. Math. Comput. 412 126595
[35] Fan J, Yin Q, Xia C, Perc M 2022 Proc. R. Soc. A. 478 20220059
 Google Scholar
Google Scholar
[36] Estrada E, Ross G J 2018 J. Theor. Biol. 438 46
 Google Scholar
Google Scholar
[37] Tudisco F, Higham D J 2021 Commun. Phys. 4 201
 Google Scholar
Google Scholar
[38] Kovalenko K, Romance M, Vasilyeva E, et al. 2022 Chaos Solitons Fractals 162 112397
 Google Scholar
Google Scholar
[39] Liu J G, Lin J H, Guo Q, Zhou T 2016 Sci. Rep. 6 21380
 Google Scholar
Google Scholar
[40] Zeng Q, Liu Y, Tang M, Gong J 2021 Knowledge-Based Syst. 229 107365
 Google Scholar
Google Scholar
[41] Li W, Nie Y, Li W, Chen X, Su S, Wang W 2022 Chaos 32 093135
 Google Scholar
Google Scholar
[42] Wang H, Ma C, Chen H S, Lai Y C, Zhang H F 2022 Nat. Commun. 13 3043
 Google Scholar
Google Scholar
[43] Génois M, Barrat A 2018 Epj Data Sci. 7 11
 Google Scholar
Google Scholar
[44] Isella L, Stehlé J, Barrat A, Cattuto C, Pinton J F, Van den Broeck W 2011 J. Theor. Biol. 271 166
 Google Scholar
Google Scholar
[45] Vanhems P, Barrat A, Cattuto C, Pinton J F, Khanafer N, Régis C, Kim B, Comte B, Voirin N 2013 PloS One 8 e73970
 Google Scholar
Google Scholar
[46] Mastrandrea R, Fournet J, Barrat A 2015 PloS One 10 e0136497
 Google Scholar
Google Scholar
Catalog
Metrics
- Abstract views: 6999
- PDF Downloads: 204
- Cited By: 0














 DownLoad:
DownLoad:




