-
In recent years, the emergence and development of single-molecule tracking technology has provided a new way to study the biological process in a cell membrane. However, the heterogeneity of biomolecular motions in cell membranes makes it difficult to distinguish and separate different molecular motion patterns from a large number of molecular trajectories. In this work, taking the system of interaction between melittin and a single-component supported phospholipid bilayer (SLB) membrane as an example, a type of data analysis method is developed through using the frequency distribution of standard deviations of single molecular motion displacements to distinguish and separate lipid molecules in different motion modes. It provides higher accuracy and more quantitative information than traditional analysis methods based on the frequency distribution of displacement or radius of gyration. By using this method, we successfully separate the two moving states of lipids in the SLB, and find that such a distribution is partly consistent with the location distribution of lipids in the upper leaflet and lower leaflet. Under the influence of surface adsorption or transmembrane poration of melittin at different concentrations, the movements of these two types of lipids are disturbed in different ways. In this work, a data analysis method for the separation of molecular motion patterns is developed according to the complex heterogeneity of molecular motions in a phospholipid membrane system. The different effects on the upper leaflet and lower leaflet of a lipid membrane due to melittin actions at different stages are obtained. The method developed in this work will be of great help in studying the dynamic processes of biological systems by using single-molecule tracking technology.
-
Keywords:
- single-molecule tracking /
- molecular dynamics /
- melittin /
- cell membrane
[1] Wimley W C 2010 ACS Chem. Biol. 5 905
 Google Scholar
Google Scholar
[2] Deng Z X, Li J L, Yuan B, Yang K 2019 Commun. Theor. Phys. 71 887
 Google Scholar
Google Scholar
[3] Yang L, Harroun T A, Weiss T M, Ding L, Huang H W 2001 Biophys. J. 81 1475
 Google Scholar
Google Scholar
[4] Wiedman G, Fuselier T, He J, Searson P C, Hristova K, Wimley W C 2014 J. Am. Chem. Soc. 136 4724
 Google Scholar
Google Scholar
[5] Liu J J, Xiao S F, Li J L, Yuan B, Yang K, Ma Y Q 2018 Biochim. Biophys. Acta, Biomembr. 1860 2234
 Google Scholar
Google Scholar
[6] Lee M T, Sun T L, Hung W C, Huang H W 2013 Proc. Natl. Acad. Sci. U. S. A. 110 14243
 Google Scholar
Google Scholar
[7] Norregaard K, Metzler R, Ritter C M, Berg-Sorensen K, Oddershede L B 2017 Chem. Rev. 117 4342
 Google Scholar
Google Scholar
[8] Shen H, Tauzin L J, Baiyasi R, Wang W X, Moringo N, Shuang B, Landes C F 2017 Chem. Rev. 117 7331
 Google Scholar
Google Scholar
[9] von Diezmann A, Shechtman Y, Moerner W E 2017 Chem. Rev. 117 7244
 Google Scholar
Google Scholar
[10] Xu C, Ma W D, Wang K, He K J, Chen Z L, Liu J J, Yang K, Yuan B 2020 J. Phys. Chem. Lett. 11 4834
 Google Scholar
Google Scholar
[11] He W, Song H, Su Y, Geng L, Ackerson B J, Peng H B, Tong P 2016 Nat. Commun. 7 11701
 Google Scholar
Google Scholar
[12] Golan Y, Sherman E 2017 Nat. Commun. 8 15851
 Google Scholar
Google Scholar
[13] Metzler R, Jeon J H, Cherstvy A G 2016 Biochim. Biophys. Acta, Biomembr. 1858 2451
 Google Scholar
Google Scholar
[14] Zwang T J, Fletcher W R, Lane T J, Johal M S 2010 Langmuir 26 4598
 Google Scholar
Google Scholar
[15] Krapf D, Campagnola G, Nepal K, Peersen O B 2016 Phys. Chem. Chem. Phys. 18 12633
 Google Scholar
Google Scholar
[16] Chen K J, Wang B, Guan J, Granick S 2013 ACS Nano 7 8634
 Google Scholar
Google Scholar
[17] Schoch R L, Barel I, Brown F L H, Haran G 2018 J. Chem. Phys. 148 123333
 Google Scholar
Google Scholar
[18] Xue C D, Shi X H, Tian Y, Zheng X, Hu G Q 2020 Nano Lett. 20 3895
 Google Scholar
Google Scholar
[19] Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A 2002 J. Cell Biol. 157 1071
 Google Scholar
Google Scholar
[20] Jeon J H, Tejedor V, Burov S, Barkai E, Selhuber-Unkel C, Berg-Sorensen K, Oddershede L, Metzler R 2011 Phys. Rev. Lett. 106 048103
 Google Scholar
Google Scholar
[21] Lubelski A, Sokolov I M, Klafter J 2008 Phys. Rev. Lett. 100 250602
 Google Scholar
Google Scholar
[22] Jeon J H, Monne H M S, Javanainen M, Metzler R 2012 Phys. Rev. Lett. 109 188103
 Google Scholar
Google Scholar
[23] Burov S, Jeon J H, Metzler R, Barkai E 2011 Phys. Chem. Chem. Phys. 13 1800
 Google Scholar
Google Scholar
-
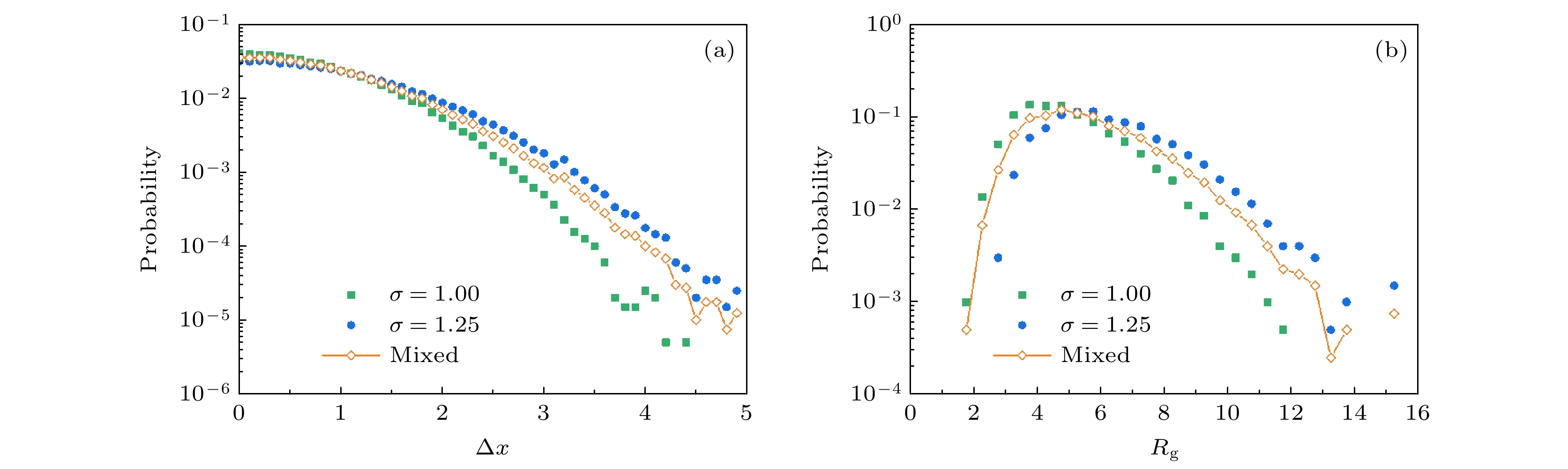
图 1 利用传统的位移和回旋半径PDF进行轨迹分析 (a) 轨迹的位移频数分布情况, 包括两类原始轨迹
$ T(\sigma = 1) $ ,$ T(\sigma = 1. 25) $ 和二者混合之后的轨迹$ {T}_{\rm{mixed}} $ ; (b) 三类轨迹的回旋半径频数分布情况Figure 1. Conventional PDF analysis of the displacement and
$ {R}_{\rm{g}} $ of trajectories: (a) Displacement PDF of the trajectories before mixing ($ T(\sigma = 1) $ ,$ T(\sigma = 1. 25) $ ) and after mixing ($ {T}_{\rm{mixed}} $ ); (b)$ {R}_{\rm{g}} $ PDF of the three conditions.图 2 利用Dstd频数分布进行轨迹分析举例 (a)
$ {T}_{\rm{mixed}}(\sigma = 1, 1. 25) $ ; (b)$ {T}_{\rm{mixed}}\left(\sigma = 1, 1. 5\right); $ (c)$ {T}_{\rm{mixed}}\left(\sigma = 1, 2\right); $ (d) 三种$ {T}_{\rm{mixed}} $ 经过Dstd轨迹分离法处理之后所得轨迹数量的准确率Figure 2. Typical examples showing trajectory analysis based on the PDF of Dstd: (a)
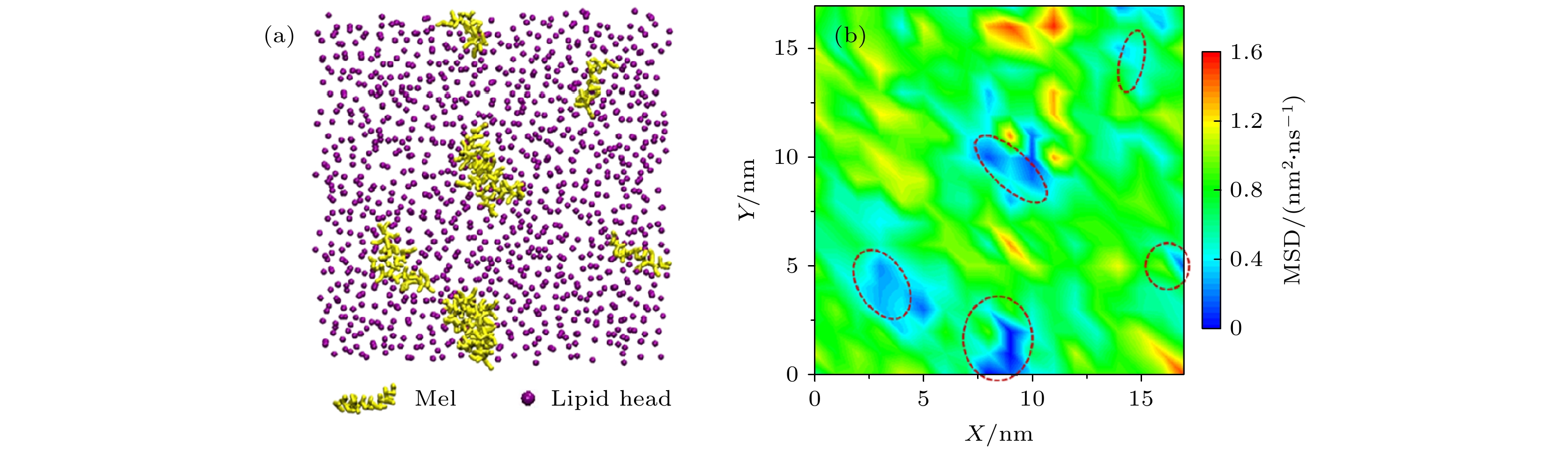
$ {T}_{\rm{mixed}}\left(\sigma = 1, 1. 25\right); $ (b)$ {T}_{\rm{mixed}}\left(\sigma = 1, 1. 5\right); $ (c)$ {T}_{\rm{mixed}}\left(\sigma = 1, 2\right); $ (d) accuracy of the seperation shown in Fig. (a) (c).图 3 DOPC支撑脂双层膜内磷脂分子运动轨迹的Dstd的频数分布分析 (a), (b) 典型的单个磷脂分子时间序列荧光成像照片(拍摄速率30 frames/s)及运动轨迹(总时长4.38 s). (c)—(e) 原始以及加入荧光淬灭剂之后的脂膜: (c)示意图; (d)分子轨迹Dstd频数分布情况; (e)
$ {T}_{\rm{slow}} $ 与$ {T}_{\rm{fast}} $ 比例柱状图. 图(c)中红色为荧光脂分子, 蓝色为无荧光脂分子Figure 3. PDF of Dstd of the lipid trajectories in a DOPC SLB before or after fluorescence quenching: (a), (b) Representative lipid molecule in a pristine membrane, its time-serial images (at 30 frames/s) and trajectory (with a time duration of 4.38 s); (c)−(e) schematic diagram, PDF of Dstd, and percentage histogram of the
$ {T}_{\rm{slow}} $ and$ {T}_{\rm{fast}} $ , of the membrane before and after outer-leaflet fluorescence quenching.图 A2 原始脂膜内脂分子运动行为的回旋半径PDF分析 (a) 不同步长下的
${R_{\rm{g}}}$ 频数分布以及相应的阈值; (b) 分别以${R_{\rm{g}}}$ 和${D_{\rm{std}}}$ 为标准分离轨迹得到的${T_{\rm{slow}}}$ 占比柱状图; (c) 纯DOPC脂膜中磷脂分子运动轨迹的回旋半径以及位移标准差之间的关系, 其中黑色线为拟合曲线, 二者呈现正相关Figure A2. PDF analysis of
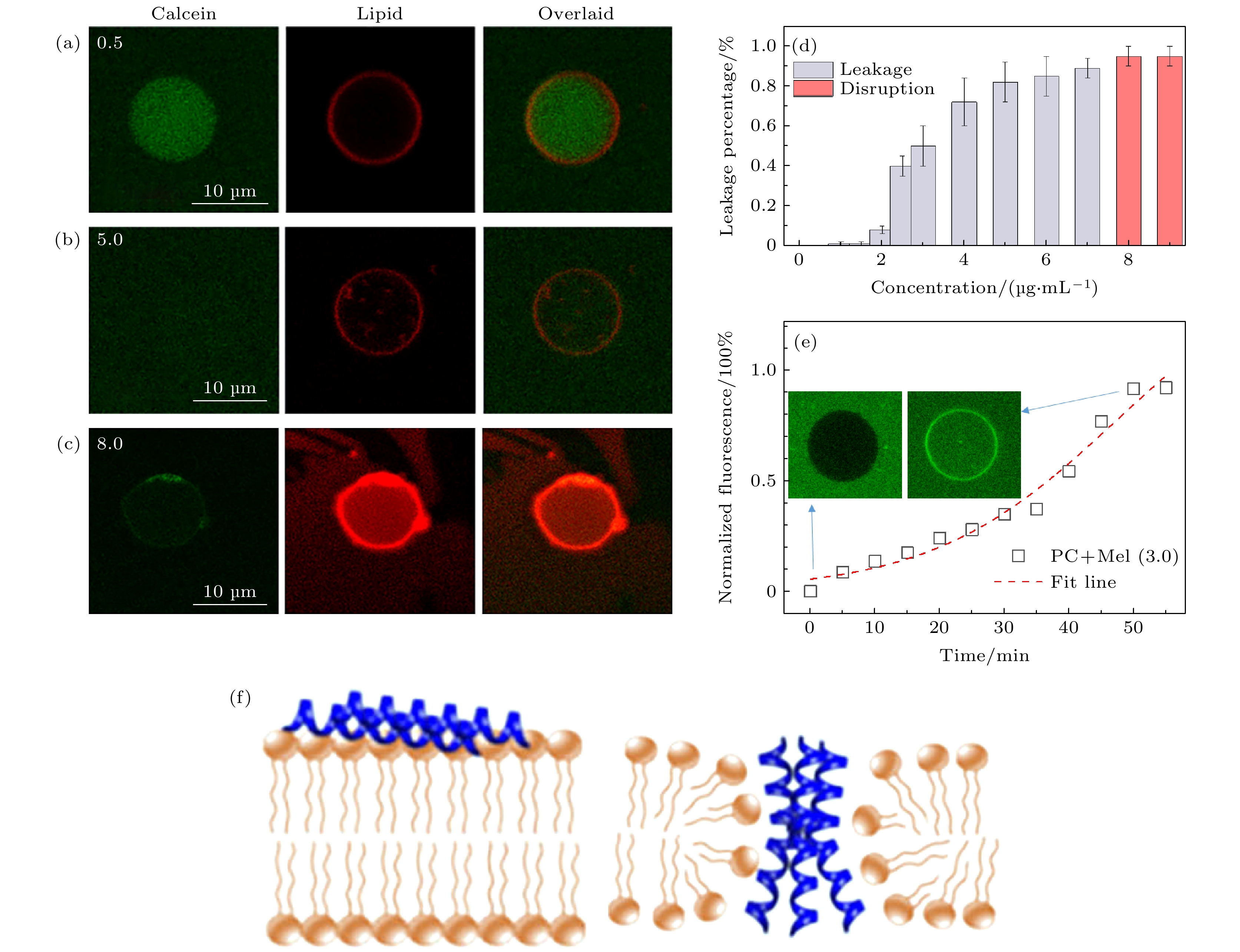
${R_{\rm{g}}}$ for the lipids in a pristine membrane: (a) PDF of${R_{\rm{g}}}$ acquried with different step values (Step = 0.1, 0.2 or 0.3). Dashed lines show determination of the threshold value separating the trajectories. (b) Percentage of${T_{\rm{slow}}}$ obtained by${R_{\rm{g}}}$ or${D_{\rm{std}}}$ analysis. (c) Relationship between${R_{\rm{g}}}$ and${D_{\rm{std}}}$ for a pure DOPC bilayer.图 A4 不同浓度的蜂毒肽作用下的GUV泄露实验 (a)—(c) 分别在0.5, 5.0以及8 μg/mL的蜂毒肽的作用下的GUV泄露情况, 其中绿色通道为GUV中包裹的钙黄绿素, 红色通道代表GUV所在的轮廓; (d) 0—10 μg/mL各浓度的蜂毒肽对GUV的扰动情况; (e)浓度3 μg/mL蜂毒肽的作用下, 典型的GUV荧光泄露曲线; (f) 低浓度蜂毒肽吸附在脂膜以及高浓度蜂毒肽形成跨膜孔道的示意图
Figure A4. GUV leakage experiment incubated with different concentration of melittin: (a)–(c) Confocal images of GUV leakage incubated with 0.5, 5.0 and 8 μg/mL of melittin; (d) influence on GUV by different concentration of melittin; (e) typical leakage curve of GUV incubated with 3 μg/mL; (f) sketch of absorption or poration of melittin interacted with membrane.
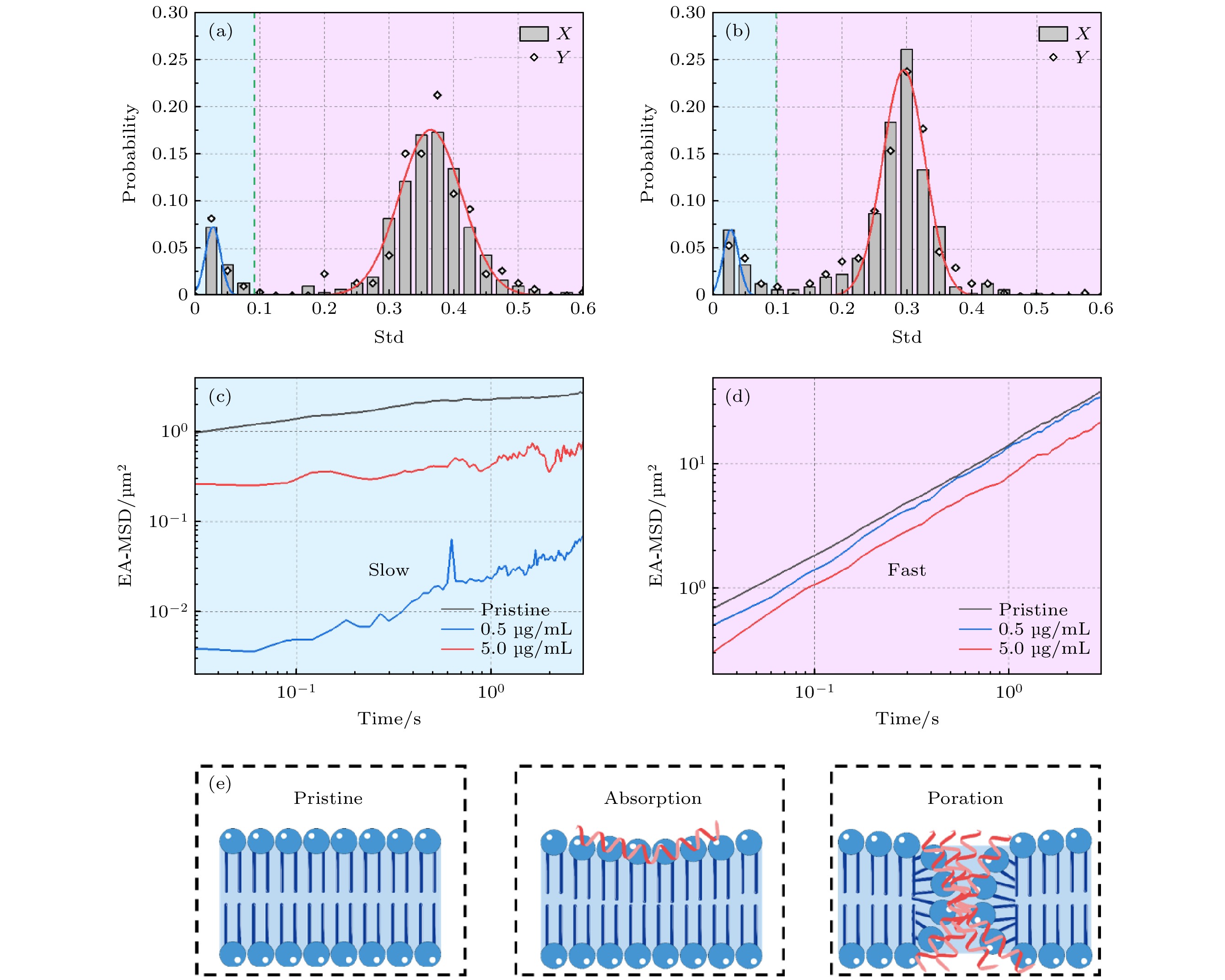
图 4 蜂毒肽作用下的脂分子运动行为分析 (a), (b) 蜂毒肽影响下磷脂分子运动轨迹的Dstd频数分布, 多肽浓度为 (a) 0.5 µg/mL, (b) 5.0 µg/mL; (c), (d) 三种实验体系内
$ {T}_{\rm{fast}} $ 与$ {T}_{\rm{slow}} $ 的系综平均均方位移; (e) 三个体系中多肽与脂膜作用示意图Figure 4. Dstd PDF analysis of the melittin-exposed lipid membrane: (a), (b) PDF of Dstd of lipids incubated with melittin at (a) 0.5 µg/mL and (b) 5.0 µg/mL; (c), (d) EA-MSD of
$ {T}_{\rm{fast}} $ and$ {T}_{\rm{slow}} $ in the three systems; (e) cartoons showing melittin-membrane interactions in the three systems.表 1 不同浓度蜂毒肽影响下磷脂膜中磷脂分子运动Dstd频数分布的拟合结果
Table 1. Fittings of the Dstd PDF of lipid trajectories in membrane incubated with melittin at different concentrations.
$ {T}_{\rm{slow}} $ $ {T}_{\rm{fast}} $ 阈值 峰位 半高宽 占比 峰位 半高宽 占比/% 原始膜 0.05 0.06 21.25% 0.37 0.11 78.75 0.16 0.5 μg/mL 0.03 0.03 12.09% 0.36 0.11 87.91 0.09 5.0 μg/mL 0.03 0.03 12.71% 0.29 0.08 87.29 0.10 R2 F-value σ = 1/σ = 1.25 0.99 24362.45 σ = 1/σ = 1.5 0.99 4661.03 σ = 1/σ = 2 0.99 5739.05 表 A2 单组分DOPC磷脂膜淬灭前后磷脂分子运动Dstd频数分布的拟合数值
Table A2. Fittings of the Dstd PDF of lipid trajectories in a SLB membrane before and after quenching.
Tslow Tfast 阈值 R2 F-value 峰位 半高宽 占比/% 峰位 半高宽 占比 原始膜 0.05 0.06 21.25 0.37 0.11 78.75 0.16 0.98 333.21 淬灭后 0.05 0.05 49.68 0.36 0.10 50.32 0.16 0.93 88.91 表 A4 不同浓度的蜂毒肽影响下磷脂分子运动轨迹的系综平均均方位移的扩散方程拟合结果
Table A4. Fittings of the EA-MSD of lipid trajectories in membrane incubated with melittin at different concentrations.
Tslow Tfast D α D α 原始膜 0.56 0.16 3.60 0.90 0.5 μg/mL 0.01 0.70 3.30 0.90 5.0 μg/mL 0.24 0.12 2.08 0.87 表 A3 不同浓度蜂毒肽影响下磷脂膜中磷脂分子运动Dstd频数分布的拟合数值
Table A3. Fittings of the Dstd PDF of lipid trajectories in membrane incubated with melittin at different concentrations.
Tslow Tfast 阈值 R2 F-value 峰位 半高宽 占比/% 峰位 半高宽 占比/% 原始膜 0.05 0.06 21.25 0.37 0.11 78.75 0.16 0.98 333.21 0.5 μg/mL 0.03 0.03 12.09 0.36 0.11 87.91 0.09 0.99 496.45 5.0 μg/mL 0.03 0.03 12.71 0.29 0.08 87.29 0.10 0.97 216.97 -
[1] Wimley W C 2010 ACS Chem. Biol. 5 905
 Google Scholar
Google Scholar
[2] Deng Z X, Li J L, Yuan B, Yang K 2019 Commun. Theor. Phys. 71 887
 Google Scholar
Google Scholar
[3] Yang L, Harroun T A, Weiss T M, Ding L, Huang H W 2001 Biophys. J. 81 1475
 Google Scholar
Google Scholar
[4] Wiedman G, Fuselier T, He J, Searson P C, Hristova K, Wimley W C 2014 J. Am. Chem. Soc. 136 4724
 Google Scholar
Google Scholar
[5] Liu J J, Xiao S F, Li J L, Yuan B, Yang K, Ma Y Q 2018 Biochim. Biophys. Acta, Biomembr. 1860 2234
 Google Scholar
Google Scholar
[6] Lee M T, Sun T L, Hung W C, Huang H W 2013 Proc. Natl. Acad. Sci. U. S. A. 110 14243
 Google Scholar
Google Scholar
[7] Norregaard K, Metzler R, Ritter C M, Berg-Sorensen K, Oddershede L B 2017 Chem. Rev. 117 4342
 Google Scholar
Google Scholar
[8] Shen H, Tauzin L J, Baiyasi R, Wang W X, Moringo N, Shuang B, Landes C F 2017 Chem. Rev. 117 7331
 Google Scholar
Google Scholar
[9] von Diezmann A, Shechtman Y, Moerner W E 2017 Chem. Rev. 117 7244
 Google Scholar
Google Scholar
[10] Xu C, Ma W D, Wang K, He K J, Chen Z L, Liu J J, Yang K, Yuan B 2020 J. Phys. Chem. Lett. 11 4834
 Google Scholar
Google Scholar
[11] He W, Song H, Su Y, Geng L, Ackerson B J, Peng H B, Tong P 2016 Nat. Commun. 7 11701
 Google Scholar
Google Scholar
[12] Golan Y, Sherman E 2017 Nat. Commun. 8 15851
 Google Scholar
Google Scholar
[13] Metzler R, Jeon J H, Cherstvy A G 2016 Biochim. Biophys. Acta, Biomembr. 1858 2451
 Google Scholar
Google Scholar
[14] Zwang T J, Fletcher W R, Lane T J, Johal M S 2010 Langmuir 26 4598
 Google Scholar
Google Scholar
[15] Krapf D, Campagnola G, Nepal K, Peersen O B 2016 Phys. Chem. Chem. Phys. 18 12633
 Google Scholar
Google Scholar
[16] Chen K J, Wang B, Guan J, Granick S 2013 ACS Nano 7 8634
 Google Scholar
Google Scholar
[17] Schoch R L, Barel I, Brown F L H, Haran G 2018 J. Chem. Phys. 148 123333
 Google Scholar
Google Scholar
[18] Xue C D, Shi X H, Tian Y, Zheng X, Hu G Q 2020 Nano Lett. 20 3895
 Google Scholar
Google Scholar
[19] Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A 2002 J. Cell Biol. 157 1071
 Google Scholar
Google Scholar
[20] Jeon J H, Tejedor V, Burov S, Barkai E, Selhuber-Unkel C, Berg-Sorensen K, Oddershede L, Metzler R 2011 Phys. Rev. Lett. 106 048103
 Google Scholar
Google Scholar
[21] Lubelski A, Sokolov I M, Klafter J 2008 Phys. Rev. Lett. 100 250602
 Google Scholar
Google Scholar
[22] Jeon J H, Monne H M S, Javanainen M, Metzler R 2012 Phys. Rev. Lett. 109 188103
 Google Scholar
Google Scholar
[23] Burov S, Jeon J H, Metzler R, Barkai E 2011 Phys. Chem. Chem. Phys. 13 1800
 Google Scholar
Google Scholar
Catalog
Metrics
- Abstract views: 6690
- PDF Downloads: 91
- Cited By: 0






















 DownLoad:
DownLoad: