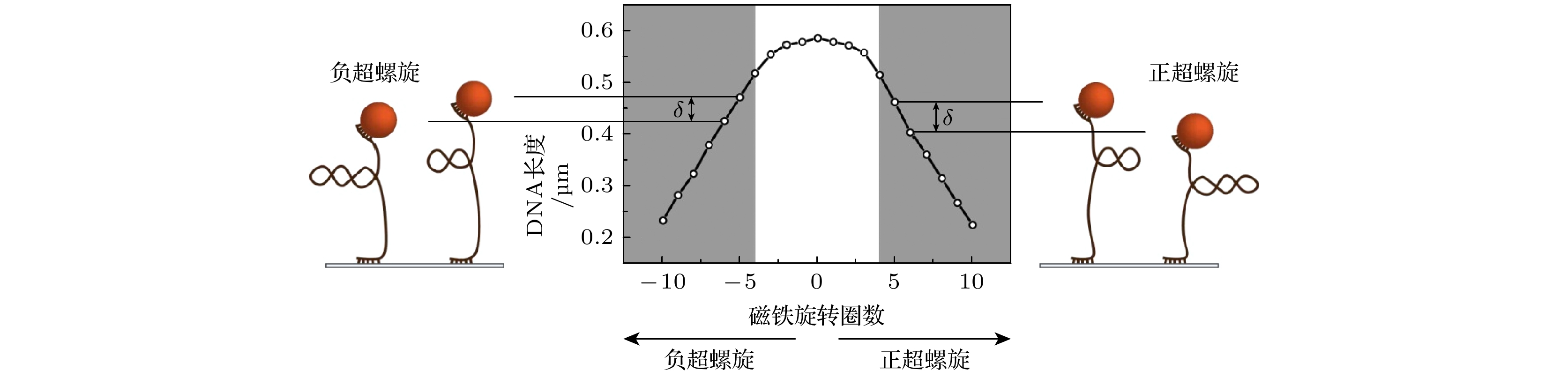
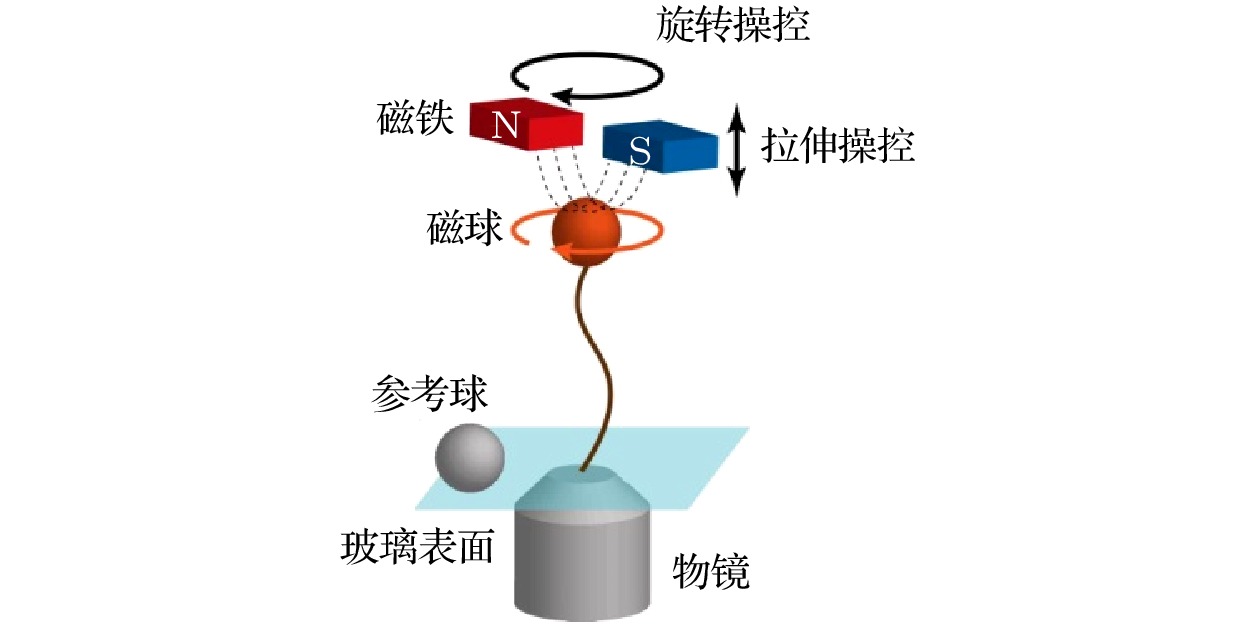
-
Gene transcription regulation is a key step for gene expression in all organisms and responsible for the transmission of genetic information and genome integrity. As one of the most important mechanisms in gene transcription, an RNA polymerase (RNAP) specifically interacts with and unwinds genome DNA to form a transcription bubble where a nascent RNA transcript is polymerized, taking one of the unwound DNA strands as its template. The RNAP translocates along the DNA to transcribe the whole gene by carrying the transcription bubble. In such a way, an RNAP completes its biological task of gene expression by physically acting as a molecular machinery. Thus, an RNAP molecule can be considered as a research object for physicists who are willing to uncover the mechanisms of life processes in a physical view. To achieve this, single-molecule method has been invented and used widely. As one of these methods, single-molecule magnetic trapping manipulates biological molecules by applying extension force or torque to the magnetic beads tethered through biological molecule to pre-coated glass surfaces by manipulating the position or rotation of a pair of magnets. A linear DNA molecule can be manipulated in such a way to generate plectonemes, i.e. DNA supercoils, under an extension force of 0.3 pN (1 pN = 10–12 N), possessing the feature that the number of unwound base pairs of a supercoiled DNA can be observed by the changes in the number of supercoils reflected by the DNA extension changes. Thus, the DNA unwound by RNAP, i.e. the transcription bubble, during transcription can be observed in this way. By monitoring the kinetics of the transcription bubble in real time, this method thus allows single-molecule detection with single-base resolution and a high-throughput data collection fashion in the kinetic studies of transcription. Owing to the advantages of the manipulation of DNA supercoils with single-molecule magnetic trapping, one can mimic the mechanistic feature of DNAs in vivo and characterize the kinetics of transcription under such conditions. This method can also be combined with single-molecule fluorescence method which can be applied to studying the mechanism of transcription regulation while monitoring the behaviors of fluorescently labeled biological molecules that interact with functional RNAP molecules, providing examples for studying the mechanisms of transcription regulations in more complex systems.
-
Keywords:
- single-molecule magnetic trapping /
- supercoiled DNA /
- gene transcription regulation /
- single-molecule kinetics
[1] Watson J D, Crick F H C 1953 Nature 171 737
 Google Scholar
Google Scholar
[2] Watson J D, Crick F H C 1953 Nature 171 964
 Google Scholar
Google Scholar
[3] Crick F 1970 Nature 227 561
 Google Scholar
Google Scholar
[4] Monod J, Changeux J P, Jacob F 1963 J. Mol. Biol. 6 306
 Google Scholar
Google Scholar
[5] Monod J, Wyman J, Changeux J P 1965 J. Mol. Biol. 12 88
 Google Scholar
Google Scholar
[6] Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A 1999 Cell 98 811
 Google Scholar
Google Scholar
[7] Gnatt A L, Cramer P, Fu J, Bushnell D A, Kornberg R D 2001 Science 292 1876
 Google Scholar
Google Scholar
[8] Chen X, Yin X, Li J, Wu Z, Qi Y, Wang X, Liu W, Xu Y 2021 Science 372 eabg0635
 Google Scholar
Google Scholar
[9] Chen X, Qi Y, Wu Z, et al. 2021 Science 372 eaba8490
 Google Scholar
Google Scholar
[10] Huang K, Wu X X, Fang C L, et al. 2021 Science 374 1579
 Google Scholar
Google Scholar
[11] Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M 2002 Mol. Cell 10 1151
 Google Scholar
Google Scholar
[12] Skourti-Stathaki K, Proudfoot N J, Gromak N 2011 Mol. Cell 42 794
 Google Scholar
Google Scholar
[13] Hazelbaker D Z, Marquardt S, Wlotzka W, Buratowski S 2013 Mol. Cell 49 55
 Google Scholar
Google Scholar
[14] Kim S, Beltran B, Irnov I, Jacobs-Wagner C 2019 Cell 179 106
 Google Scholar
Google Scholar
[15] Abbondanzieri E A, Greenleaf W J, Shaevitz J W, Landick R, Block S M 2005 Nature 438 460
 Google Scholar
Google Scholar
[16] Chakraborty A, Wang D, Ebright Y W, et al. 2012 Science 337 591
 Google Scholar
Google Scholar
[17] Howan K, Smith A J, Westblade L F, Joly N, Grange W, Zorman S, Darst S A, Savery N J, Strick T R 2012 Nature 490 431
 Google Scholar
Google Scholar
[18] Ma J, Bai L, Wang M D 2013 Science 340 1580
 Google Scholar
Google Scholar
[19] Koh H R, Roy R, Sorokina M, Tang G Q, Nandakumar D, Patel S S, Ha T 2018 Mol. Cell 70 695
 Google Scholar
Google Scholar
[20] Rosen G A, Baek I, Friedman L J, Joo Y J, Buratowski S, Gelles J 2020 Proc. Natl. Acad. Sci. 117 32348
 Google Scholar
Google Scholar
[21] Neuman K C, Nagy A 2008 Nat. Methods 5 491
 Google Scholar
Google Scholar
[22] Roy R, Hohng S, Ha T J 2008 Nat. Methods 5 507
 Google Scholar
Google Scholar
[23] Gosse C, Croquette V 2002 Biophys. J. 82 3314
 Google Scholar
Google Scholar
[24] Cnossen J P, Dulin D, Dekker N H 2014 Rev. Sci. Instrum. 85 103712
 Google Scholar
Google Scholar
[25] Huhle A, Klaue D, Brutzer H, Daldrop P, Joo S, Otto O, Keyser U F, Seidel R 2015 Nat. Commun. 6 5885
 Google Scholar
Google Scholar
[26] Ashkin A, Dziedzic J M, Bjorkholm J E, Chu S 1986 Opt. Lett. 11 288
 Google Scholar
Google Scholar
[27] Ashkin A, Dziedzic J M 1987 Science 235 1517
 Google Scholar
Google Scholar
[28] Neuman K C, Block S M 2004 Rev. Sci. Instrum. 75 2787
 Google Scholar
Google Scholar
[29] Rief M, Gautel M, Oesterhelt F, Femandez J M, Gaub H E 1997 Science 276 1109
 Google Scholar
Google Scholar
[30] Yu H, Siewny M G W, Edwards D T, Sanders A W, Perkins T T 2017 Science 355 945
 Google Scholar
Google Scholar
[31] Jiao F, Cannon K S, Lin Y C, Gladfelter A S, Scheuring S 2020 Nat. Commu. 11 5062
 Google Scholar
Google Scholar
[32] Ha T, Enderle T, Ogletree D F, Chemla D S, Selvin P R, Weiss S 1996 Proc. Natl. Acad. Sci. 93 6264
 Google Scholar
Google Scholar
[33] Weiss S 1999 Science 283 1676
 Google Scholar
Google Scholar
[34] Lerner E, Cordes T, Ingargiola A, Alhadid Y, Chung S, Michalet X, Weiss S 2018 Science 359 eaan1133
 Google Scholar
Google Scholar
[35] Friedman L J, Chung J, Gelles J 2006 Biophys. J. 91 1023
 Google Scholar
Google Scholar
[36] Friedman L J, Gelles J 2015 Methods 86 27
 Google Scholar
Google Scholar
[37] Thompson M A, Lew M D, Moerner W 2012 Annu. Rev. Bioph. Biom. 41 321
 Google Scholar
Google Scholar
[38] Betzig E, Patterson G H, Sougrat R, Lindwasser O W, Olenych S, Bonifacino J S, Davidson M W, Lippincott-Schwartz J, Hess H F 2006 Science 313 1642
 Google Scholar
Google Scholar
[39] Bates M, Huang B, Dempsey G T, Zhuang X 2007 Science 317 1749
 Google Scholar
Google Scholar
[40] Seol Y, Neuman K C 2017 Combined Magnetic Tweezers, Micro-mirror Total Internal Reflection Fluorescence Microscope for Single-molecule Manipulation and Visualization Single Molecule Analysis (New York: Springer) pp297–316
[41] Fan J, Leroux-Coyau M, Savery N J, Strick T R 2016 Nature 536 234
 Google Scholar
Google Scholar
[42] Comstock M J, Whitley K D, Jia H, Sokoloski J, Lohman T M, Ha T, Chemla Y R 2015 Science 348 352
 Google Scholar
Google Scholar
[43] Lionnet T, Allemand J F, Revyakin A, Strick T R, Saleh O A, Bensimon D, Croquette V 2011 Cold Spring Harb. Protoc. 2012 133
 Google Scholar
Google Scholar
[44] Smith S B, Finzi L, Bustamante C 1992 Science 258 1122
 Google Scholar
Google Scholar
[45] Strick T R, Allemand J F, Bensimon D, Bensimon A, Croquette V 1996 Science 271 1835
 Google Scholar
Google Scholar
[46] Chen H, Yuan G, Winardhi R S, Yao M, Popa I, Fernandez J M, Yan J 2015 J. Am. Chem. Soc. 137 3540
 Google Scholar
Google Scholar
[47] Guo Z, Hong H, Yuan G, Qian H, Li B, Cao Y, Wang W, Wu C X, Chen H 2020 Phys. Rev. Lett. 125 198101
 Google Scholar
Google Scholar
[48] Strick T, Allemand J F, Bensimon D, Croquette V 1998 Biophys. J. 74 2016
 Google Scholar
Google Scholar
[49] Revyakin A, Ebright R H, Strick T R 2004 Proc. Natl. Acad. Sci. U. S. A. 101 4776
 Google Scholar
Google Scholar
[50] Revyakin A, Ebright R H, Strick T R 2005 Nat. Methods 2 127
 Google Scholar
Google Scholar
[51] Yu L, Winkelman J T, Pukhrambam C, Strick T R, Nickels B E, Ebright R H 2017 eLife 6 e32038
 Google Scholar
Google Scholar
[52] Smith S B, Cui Y, Bustamante C 1996 Science 271 795
 Google Scholar
Google Scholar
[53] Roeder R G 2019 Nat Struct. Mol. Biol. 26 783
 Google Scholar
Google Scholar
[54] Pomerantz R T, O’Donnell M 2010 Science 327 590
 Google Scholar
Google Scholar
[55] Merrikh H, Machón C, Grainger W H, Grossman A D, Soultanas P 2011 Nature 470 554
 Google Scholar
Google Scholar
[56] Tomko E J, Fishburn J, Hahn S, Galburt E A 2017 Nat Struct. Mol. Biol. 24 1139
 Google Scholar
Google Scholar
[57] Revyakin A, Liu C, Ebright R H, Strick T R 2006 Science 314 1139
 Google Scholar
Google Scholar
[58] Kapanidis A N, Margeat E, Ho S O, Kortkhonjia E, Weiss S, Ebright R H 2006 Science 314 1144
 Google Scholar
Google Scholar
[59] Lerner E, Chung S, Allen B L, et al. 2016 Proc. Natl. Acad. Sci. U. S. A. 113 6562
 Google Scholar
Google Scholar
[60] Wang D, Bushnell D A, Huang X, Westover K D, Levitt M, Kornberg R D 2009 Science 324 1203
 Google Scholar
Google Scholar
[61] Zhang Y, Han W, Wang L, Wang H, Jia Q, Chen T, Wang S, Li M 2023 J. Phys. Chem. B 127 2909
 Google Scholar
Google Scholar
[62] Zhu C, Guo X, Dumas P, Takacs M, Abdelkareem M, Broeck A V, Saint-André C, Papai G, Crucifix C, Ortiz J, Weixlbaumer A 2022 Nat. Commun. 13 1546
 Google Scholar
Google Scholar
[63] Ray-Soni A, Bellecourt M J, Landick R 2016 Annu. Rev. Biochem. 85 319
 Google Scholar
Google Scholar
[64] Kim M, Vasiljeva L, Rando O J, Zhelkovsky A, Moore C, Buratowski S 2006 Mol. Cell 24 723
 Google Scholar
Google Scholar
[65] Arndt K M, Reines D 2015 Annu. Rev. Biochem. 84 381
 Google Scholar
Google Scholar
[66] Fazal F M, Meng C A, Murakami K, Kornberg R D, Block S M 2015 Nature 525 274
 Google Scholar
Google Scholar
[67] Wang S, Han Z, Libri D, Porrua O, Strick T R 2019 Nat. Commun. 10 1545
 Google Scholar
Google Scholar
[68] Klein H L, Ang K K, Arkin M R, et al. 2019 Microbial. Cell 6 65
 Google Scholar
Google Scholar
[69] 王爽, 郑海子, 赵振业, 陆越, 徐春华 2013 62 168703
 Google Scholar
Google Scholar
Wang S, Zheng H Z, Zhao Z Y, Lu Y, Xu C H 2013 Acta Phys. Sin. 62 168703
 Google Scholar
Google Scholar
[70] Graves E T, Duboc C, Fan J, Stransky F, Leroux-Coyau M, Strick T R 2015 Nat. Struct. Mol. Biol. 22 452
 Google Scholar
Google Scholar
-
图 4 单分子磁镊操控技术和单分子荧光成像技术联用方案 (a)方案示意图; (b)磁镊监测转录动力学过程; (c)单分子荧光方法同步观测RNAP、Mfd和RNA的时空分布[68]
Figure 4. Schematic for combination of single-molecule magnetic trap and single-molecule fluorescence imaging: (a) Schematic of the assay; (b) transcription kinetics characterized via single-molecule magnetic trap; (c) simultaneous detection of RNAP, Mfd and RNA via single-molecule fluorescence assay[68].
-
[1] Watson J D, Crick F H C 1953 Nature 171 737
 Google Scholar
Google Scholar
[2] Watson J D, Crick F H C 1953 Nature 171 964
 Google Scholar
Google Scholar
[3] Crick F 1970 Nature 227 561
 Google Scholar
Google Scholar
[4] Monod J, Changeux J P, Jacob F 1963 J. Mol. Biol. 6 306
 Google Scholar
Google Scholar
[5] Monod J, Wyman J, Changeux J P 1965 J. Mol. Biol. 12 88
 Google Scholar
Google Scholar
[6] Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A 1999 Cell 98 811
 Google Scholar
Google Scholar
[7] Gnatt A L, Cramer P, Fu J, Bushnell D A, Kornberg R D 2001 Science 292 1876
 Google Scholar
Google Scholar
[8] Chen X, Yin X, Li J, Wu Z, Qi Y, Wang X, Liu W, Xu Y 2021 Science 372 eabg0635
 Google Scholar
Google Scholar
[9] Chen X, Qi Y, Wu Z, et al. 2021 Science 372 eaba8490
 Google Scholar
Google Scholar
[10] Huang K, Wu X X, Fang C L, et al. 2021 Science 374 1579
 Google Scholar
Google Scholar
[11] Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M 2002 Mol. Cell 10 1151
 Google Scholar
Google Scholar
[12] Skourti-Stathaki K, Proudfoot N J, Gromak N 2011 Mol. Cell 42 794
 Google Scholar
Google Scholar
[13] Hazelbaker D Z, Marquardt S, Wlotzka W, Buratowski S 2013 Mol. Cell 49 55
 Google Scholar
Google Scholar
[14] Kim S, Beltran B, Irnov I, Jacobs-Wagner C 2019 Cell 179 106
 Google Scholar
Google Scholar
[15] Abbondanzieri E A, Greenleaf W J, Shaevitz J W, Landick R, Block S M 2005 Nature 438 460
 Google Scholar
Google Scholar
[16] Chakraborty A, Wang D, Ebright Y W, et al. 2012 Science 337 591
 Google Scholar
Google Scholar
[17] Howan K, Smith A J, Westblade L F, Joly N, Grange W, Zorman S, Darst S A, Savery N J, Strick T R 2012 Nature 490 431
 Google Scholar
Google Scholar
[18] Ma J, Bai L, Wang M D 2013 Science 340 1580
 Google Scholar
Google Scholar
[19] Koh H R, Roy R, Sorokina M, Tang G Q, Nandakumar D, Patel S S, Ha T 2018 Mol. Cell 70 695
 Google Scholar
Google Scholar
[20] Rosen G A, Baek I, Friedman L J, Joo Y J, Buratowski S, Gelles J 2020 Proc. Natl. Acad. Sci. 117 32348
 Google Scholar
Google Scholar
[21] Neuman K C, Nagy A 2008 Nat. Methods 5 491
 Google Scholar
Google Scholar
[22] Roy R, Hohng S, Ha T J 2008 Nat. Methods 5 507
 Google Scholar
Google Scholar
[23] Gosse C, Croquette V 2002 Biophys. J. 82 3314
 Google Scholar
Google Scholar
[24] Cnossen J P, Dulin D, Dekker N H 2014 Rev. Sci. Instrum. 85 103712
 Google Scholar
Google Scholar
[25] Huhle A, Klaue D, Brutzer H, Daldrop P, Joo S, Otto O, Keyser U F, Seidel R 2015 Nat. Commun. 6 5885
 Google Scholar
Google Scholar
[26] Ashkin A, Dziedzic J M, Bjorkholm J E, Chu S 1986 Opt. Lett. 11 288
 Google Scholar
Google Scholar
[27] Ashkin A, Dziedzic J M 1987 Science 235 1517
 Google Scholar
Google Scholar
[28] Neuman K C, Block S M 2004 Rev. Sci. Instrum. 75 2787
 Google Scholar
Google Scholar
[29] Rief M, Gautel M, Oesterhelt F, Femandez J M, Gaub H E 1997 Science 276 1109
 Google Scholar
Google Scholar
[30] Yu H, Siewny M G W, Edwards D T, Sanders A W, Perkins T T 2017 Science 355 945
 Google Scholar
Google Scholar
[31] Jiao F, Cannon K S, Lin Y C, Gladfelter A S, Scheuring S 2020 Nat. Commu. 11 5062
 Google Scholar
Google Scholar
[32] Ha T, Enderle T, Ogletree D F, Chemla D S, Selvin P R, Weiss S 1996 Proc. Natl. Acad. Sci. 93 6264
 Google Scholar
Google Scholar
[33] Weiss S 1999 Science 283 1676
 Google Scholar
Google Scholar
[34] Lerner E, Cordes T, Ingargiola A, Alhadid Y, Chung S, Michalet X, Weiss S 2018 Science 359 eaan1133
 Google Scholar
Google Scholar
[35] Friedman L J, Chung J, Gelles J 2006 Biophys. J. 91 1023
 Google Scholar
Google Scholar
[36] Friedman L J, Gelles J 2015 Methods 86 27
 Google Scholar
Google Scholar
[37] Thompson M A, Lew M D, Moerner W 2012 Annu. Rev. Bioph. Biom. 41 321
 Google Scholar
Google Scholar
[38] Betzig E, Patterson G H, Sougrat R, Lindwasser O W, Olenych S, Bonifacino J S, Davidson M W, Lippincott-Schwartz J, Hess H F 2006 Science 313 1642
 Google Scholar
Google Scholar
[39] Bates M, Huang B, Dempsey G T, Zhuang X 2007 Science 317 1749
 Google Scholar
Google Scholar
[40] Seol Y, Neuman K C 2017 Combined Magnetic Tweezers, Micro-mirror Total Internal Reflection Fluorescence Microscope for Single-molecule Manipulation and Visualization Single Molecule Analysis (New York: Springer) pp297–316
[41] Fan J, Leroux-Coyau M, Savery N J, Strick T R 2016 Nature 536 234
 Google Scholar
Google Scholar
[42] Comstock M J, Whitley K D, Jia H, Sokoloski J, Lohman T M, Ha T, Chemla Y R 2015 Science 348 352
 Google Scholar
Google Scholar
[43] Lionnet T, Allemand J F, Revyakin A, Strick T R, Saleh O A, Bensimon D, Croquette V 2011 Cold Spring Harb. Protoc. 2012 133
 Google Scholar
Google Scholar
[44] Smith S B, Finzi L, Bustamante C 1992 Science 258 1122
 Google Scholar
Google Scholar
[45] Strick T R, Allemand J F, Bensimon D, Bensimon A, Croquette V 1996 Science 271 1835
 Google Scholar
Google Scholar
[46] Chen H, Yuan G, Winardhi R S, Yao M, Popa I, Fernandez J M, Yan J 2015 J. Am. Chem. Soc. 137 3540
 Google Scholar
Google Scholar
[47] Guo Z, Hong H, Yuan G, Qian H, Li B, Cao Y, Wang W, Wu C X, Chen H 2020 Phys. Rev. Lett. 125 198101
 Google Scholar
Google Scholar
[48] Strick T, Allemand J F, Bensimon D, Croquette V 1998 Biophys. J. 74 2016
 Google Scholar
Google Scholar
[49] Revyakin A, Ebright R H, Strick T R 2004 Proc. Natl. Acad. Sci. U. S. A. 101 4776
 Google Scholar
Google Scholar
[50] Revyakin A, Ebright R H, Strick T R 2005 Nat. Methods 2 127
 Google Scholar
Google Scholar
[51] Yu L, Winkelman J T, Pukhrambam C, Strick T R, Nickels B E, Ebright R H 2017 eLife 6 e32038
 Google Scholar
Google Scholar
[52] Smith S B, Cui Y, Bustamante C 1996 Science 271 795
 Google Scholar
Google Scholar
[53] Roeder R G 2019 Nat Struct. Mol. Biol. 26 783
 Google Scholar
Google Scholar
[54] Pomerantz R T, O’Donnell M 2010 Science 327 590
 Google Scholar
Google Scholar
[55] Merrikh H, Machón C, Grainger W H, Grossman A D, Soultanas P 2011 Nature 470 554
 Google Scholar
Google Scholar
[56] Tomko E J, Fishburn J, Hahn S, Galburt E A 2017 Nat Struct. Mol. Biol. 24 1139
 Google Scholar
Google Scholar
[57] Revyakin A, Liu C, Ebright R H, Strick T R 2006 Science 314 1139
 Google Scholar
Google Scholar
[58] Kapanidis A N, Margeat E, Ho S O, Kortkhonjia E, Weiss S, Ebright R H 2006 Science 314 1144
 Google Scholar
Google Scholar
[59] Lerner E, Chung S, Allen B L, et al. 2016 Proc. Natl. Acad. Sci. U. S. A. 113 6562
 Google Scholar
Google Scholar
[60] Wang D, Bushnell D A, Huang X, Westover K D, Levitt M, Kornberg R D 2009 Science 324 1203
 Google Scholar
Google Scholar
[61] Zhang Y, Han W, Wang L, Wang H, Jia Q, Chen T, Wang S, Li M 2023 J. Phys. Chem. B 127 2909
 Google Scholar
Google Scholar
[62] Zhu C, Guo X, Dumas P, Takacs M, Abdelkareem M, Broeck A V, Saint-André C, Papai G, Crucifix C, Ortiz J, Weixlbaumer A 2022 Nat. Commun. 13 1546
 Google Scholar
Google Scholar
[63] Ray-Soni A, Bellecourt M J, Landick R 2016 Annu. Rev. Biochem. 85 319
 Google Scholar
Google Scholar
[64] Kim M, Vasiljeva L, Rando O J, Zhelkovsky A, Moore C, Buratowski S 2006 Mol. Cell 24 723
 Google Scholar
Google Scholar
[65] Arndt K M, Reines D 2015 Annu. Rev. Biochem. 84 381
 Google Scholar
Google Scholar
[66] Fazal F M, Meng C A, Murakami K, Kornberg R D, Block S M 2015 Nature 525 274
 Google Scholar
Google Scholar
[67] Wang S, Han Z, Libri D, Porrua O, Strick T R 2019 Nat. Commun. 10 1545
 Google Scholar
Google Scholar
[68] Klein H L, Ang K K, Arkin M R, et al. 2019 Microbial. Cell 6 65
 Google Scholar
Google Scholar
[69] 王爽, 郑海子, 赵振业, 陆越, 徐春华 2013 62 168703
 Google Scholar
Google Scholar
Wang S, Zheng H Z, Zhao Z Y, Lu Y, Xu C H 2013 Acta Phys. Sin. 62 168703
 Google Scholar
Google Scholar
[70] Graves E T, Duboc C, Fan J, Stransky F, Leroux-Coyau M, Strick T R 2015 Nat. Struct. Mol. Biol. 22 452
 Google Scholar
Google Scholar
Catalog
Metrics
- Abstract views: 4983
- PDF Downloads: 154
- Cited By: 0














 DownLoad:
DownLoad: