-
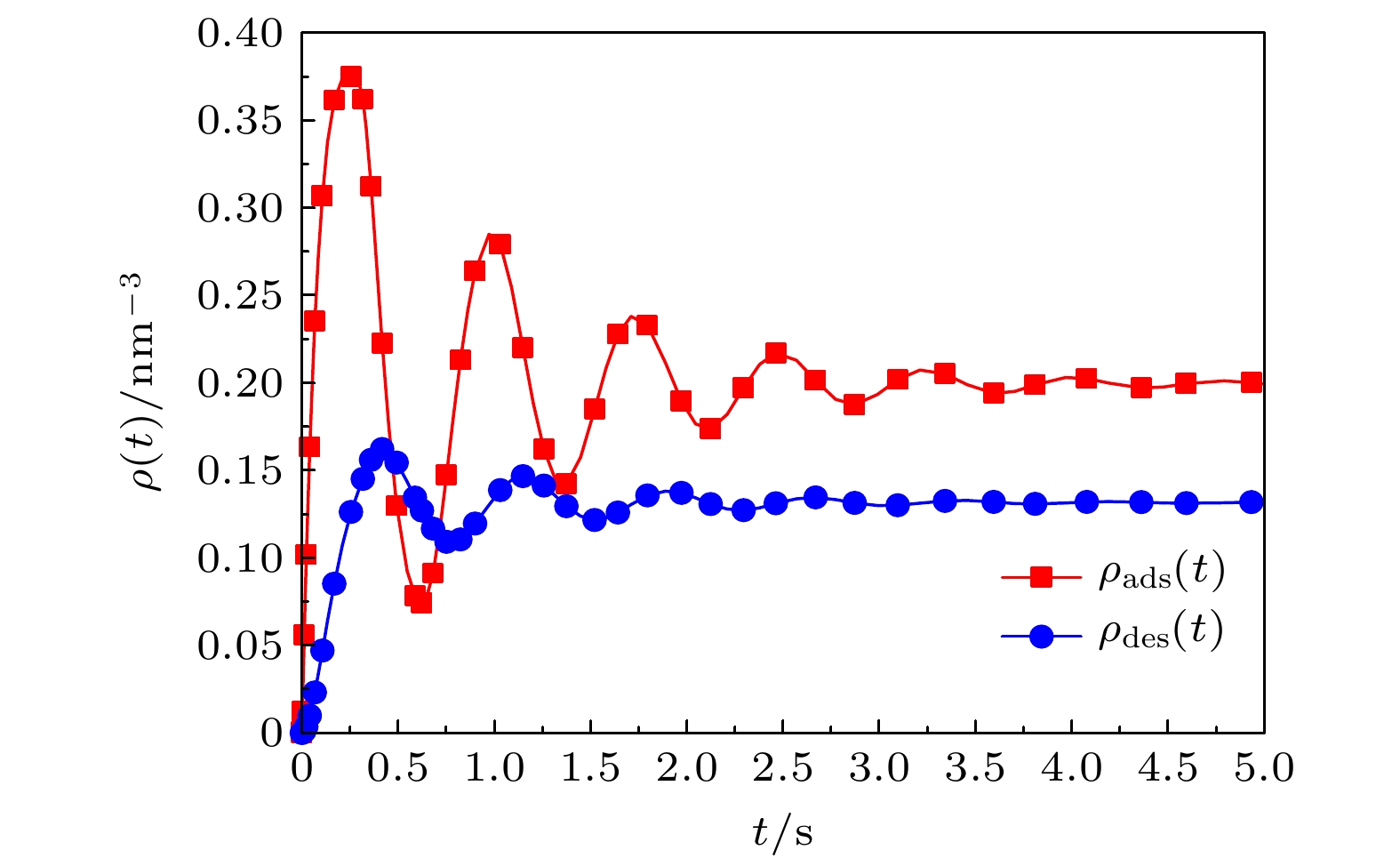
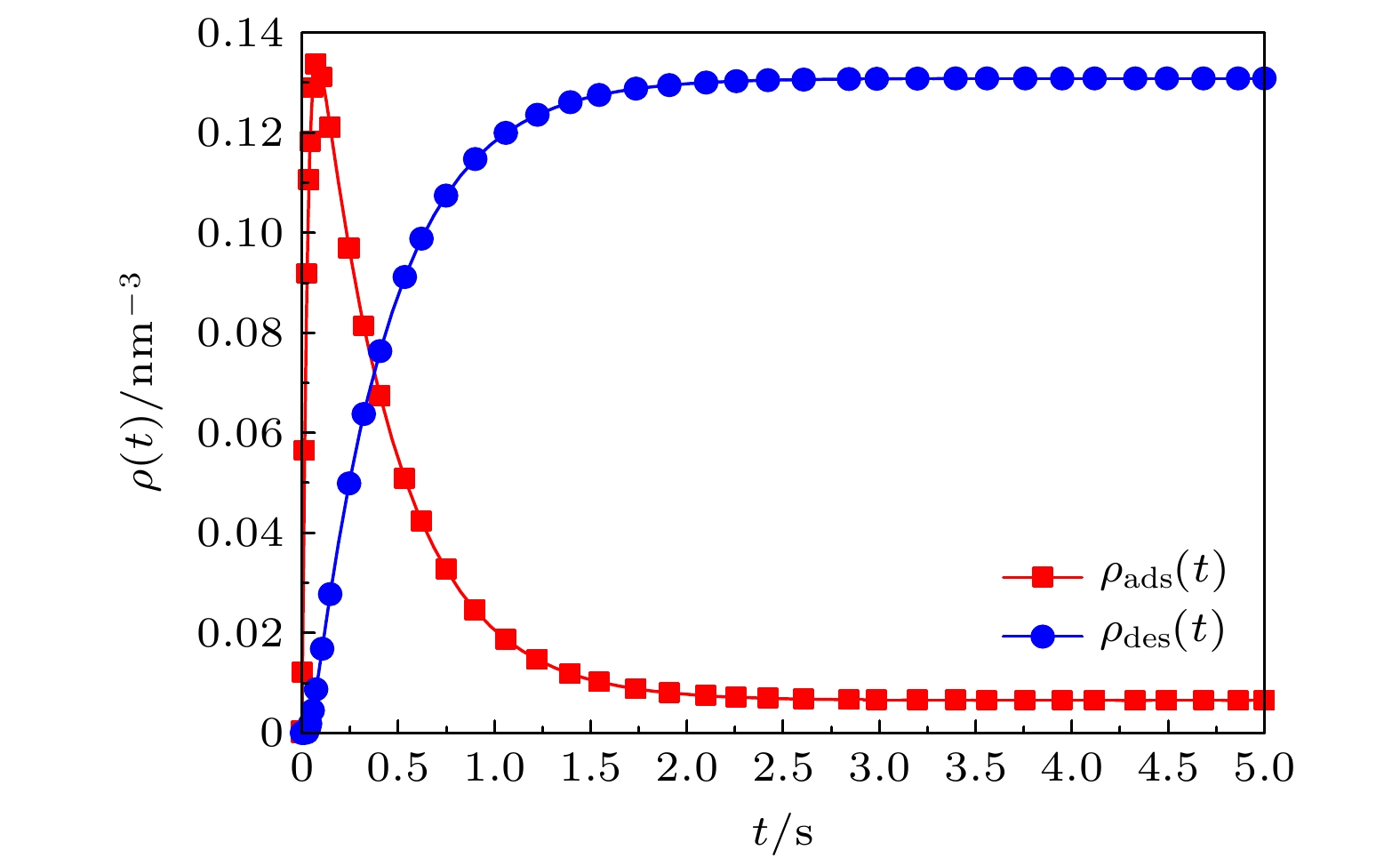
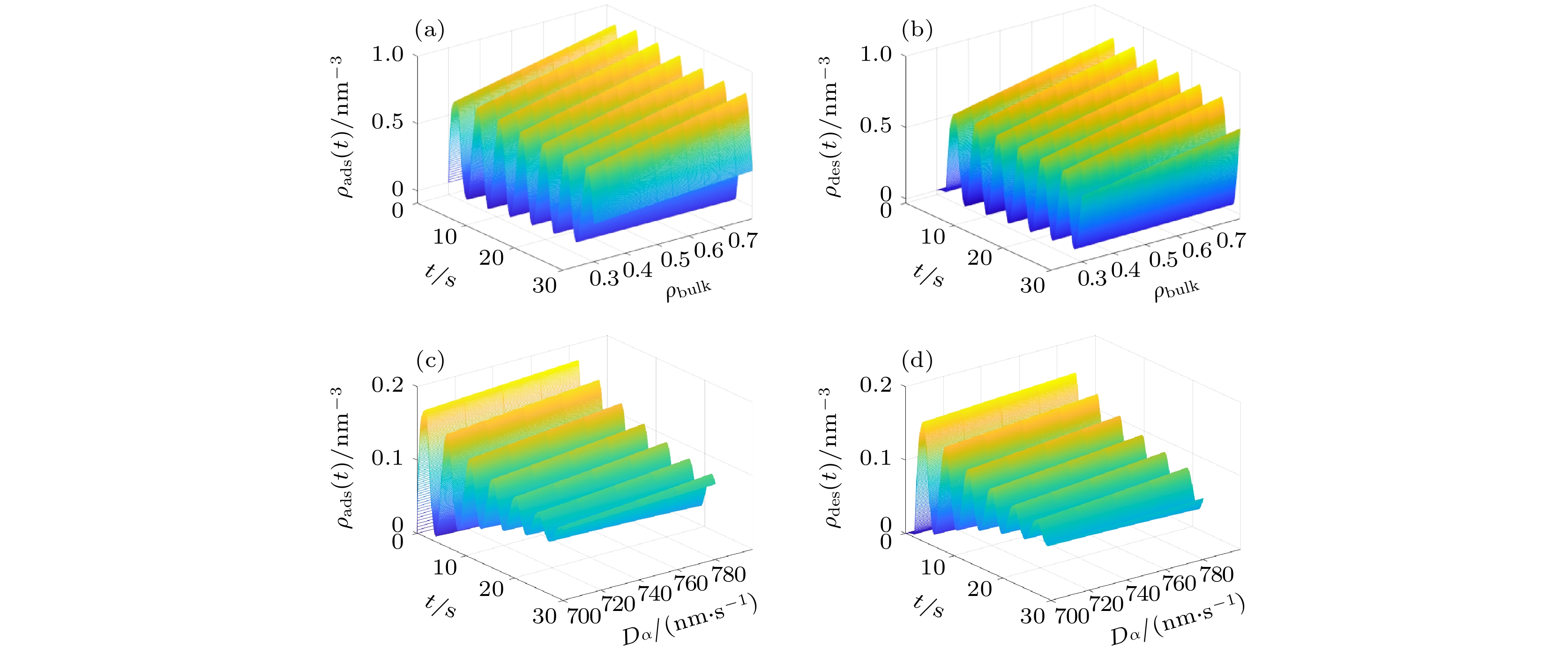
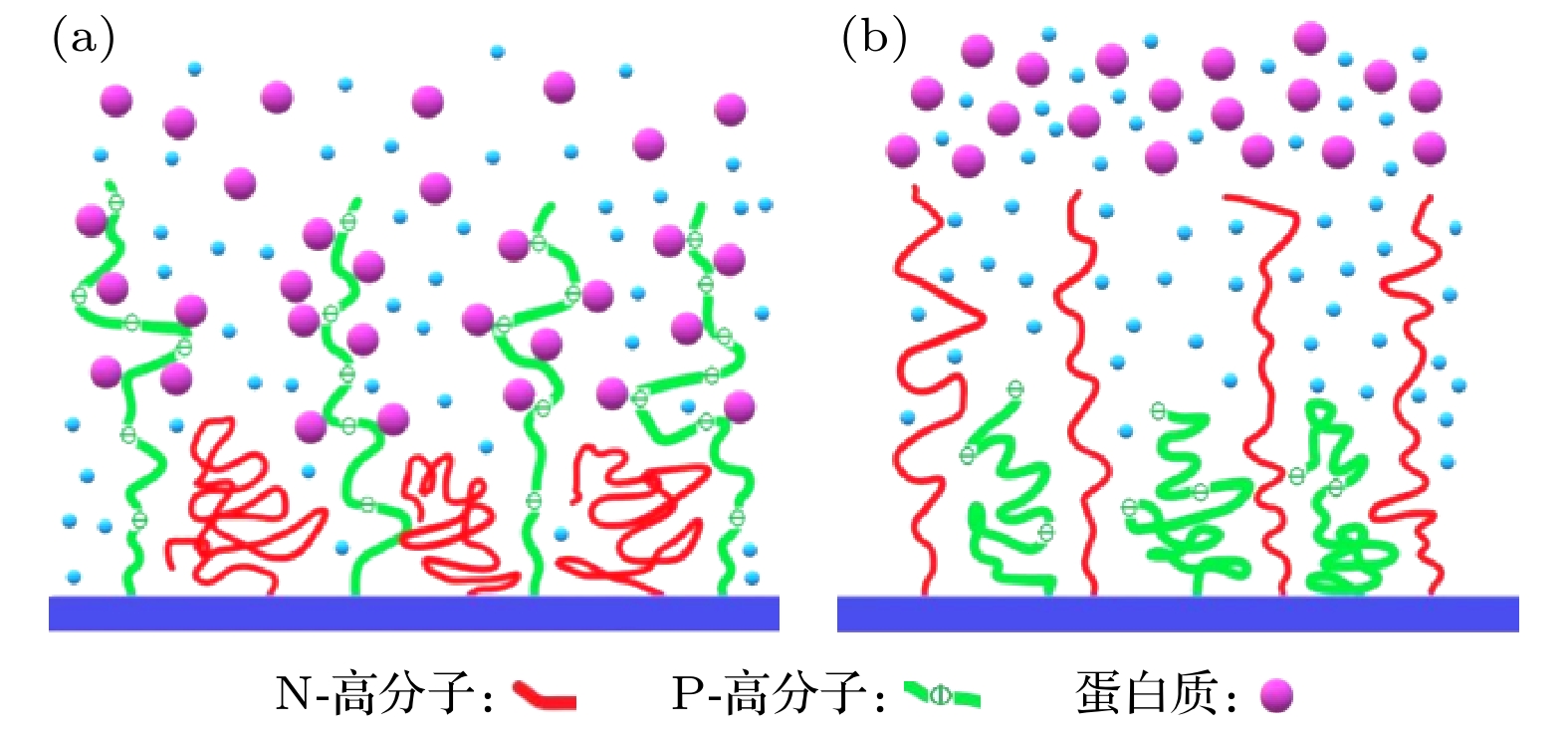
Based on the diffusion dynamics, a new theoretical model is established to investigate the dynamic properties of a polymer mixed brush (PMB) in the protein adsorption/desorption process. The theoretical model considers the adsorption of proteins by one polymer chain (P-polymer chain) and the desorption of proteins by another polymer chain (N-polymer chain) in a PMB, as well as the time delay between adsorption and desorption. The dynamic properties of protein adsorption/desorption by a PMB depend on not only the chemical and physical properties of polymer chains, but also the microenvironment (density of protein in the solution and protein diffusivity) of the PMB. In order to describe the different chemical and physical properties of polymer chains and microenvironments in PMB, we take different model parameters, and obtain partial adsorption/desorption, complete adsorption/desorption and periodic adsorption/desorption of proteins by the PMB. By analyzing the process of protein adsorption/desorption in a PMB, we find that the microenvironment has an obvious influence on the adsorption and desorption of protein by the PMB. It is also shown that the adsorption of protein and the desorption of protein by the PMB have a stable and invariable periodic cycle when an alternating electric field is applied. The average adsorption capacity and the average desorption capacity increase in comparison with those when no electric field is applied. A stable alternating electric field enables the PMB to exhibit stable periodic dynamic characteristics in the dynamic process of protein adsorption and desorption. Our theoretical results are consistent with the experimental observations. Based on this, it is predicted that an external electric field can realize multiple cycles of protein adsorption and desorption by PMB, which provides necessary references and useful insights into controllable protein adsorption/desorption by the PMB in the practical applications.
-
Keywords:
- polymer mixed brush /
- protein /
- adsorption/desorption /
- dynamic properties
[1] Tan J S, Martic P A 1990 J. Colloid Interf. Sci. 136 415
 Google Scholar
Google Scholar
[2] Steadman B L, Thompson K C, Middaugh C R, Matsuno K, Vrona S, Lawson E Q, Lewis R V 1992 Biotechn. Bioengin. 40 8
 Google Scholar
Google Scholar
[3] Norde W, Gage D 2004 Langmuir 20 4162
 Google Scholar
Google Scholar
[4] Moerz S T, Huber P 2015 J. Phys. Chem. C 119 27072
 Google Scholar
Google Scholar
[5] Ladam G, Gergely C, Senger B, Decher G, Voegel J C, Schaaf P, Cuisinier F J 2000 Biomacromolecules 1 674
 Google Scholar
Google Scholar
[6] Wittemann A, Ballauff M 2006 Phys. Chem. Chem. Phys. 8 5269
 Google Scholar
Google Scholar
[7] Yang J, Hua Z, Wang T, Wu B, Liu G, Zhang G Z 2015 Langmuir 31 6078
 Google Scholar
Google Scholar
[8] Delcroix M F, Huet G L, Conard T, Demoustier-Champagne S, Prez F E D, Landoulsi J, Dupont-Gillain C C 2013 Biomacromolecules 14 215
 Google Scholar
Google Scholar
[9] Bratek-Skicki A, Cristaudo V, Savocco J, et al. 2019 Biomacromolecules 20 778
 Google Scholar
Google Scholar
[10] Pan C, Liu X R, Gong K, Mumtaz F, Wang Y M 2018 J. Mater. Chem. B 6 556
[11] Halperin A 1999 Langmuir 15 2525
 Google Scholar
Google Scholar
[12] Fang F, Satulovsky J, Szleifer I 2005 Biophys. J. 89 1516
 Google Scholar
Google Scholar
[13] Su X H, Lei Q L, Ren C L 2015 Chin. Phys. B 24 113601
 Google Scholar
Google Scholar
[14] Biesheuvel P M, Wittemann A 2005 J. Phys. Chem. B 109 4209
[15] Biesheuvel P M, Leermakers F, Stuart M 2006 Phys. Rev. E 73 011802
[16] Xu Y L, Chen X Q, Chen H Y, Xu S H, Liu H L, Hu Y 2012 Mol. Simulat. 38 274
 Google Scholar
Google Scholar
[17] Szleifer I, Carignano M A 2000 Macromol. Rapid Commun. 21 423
 Google Scholar
Google Scholar
[18] Fang F, Szleifer I 2001 Biophys. J. 80 2568
 Google Scholar
Google Scholar
[19] Fang F, Szleifer I 2002 Langmuir 18 5497
 Google Scholar
Google Scholar
[20] Weir M P, Heriot S Y, Martin S J, Parnell A J, Holt S A, Webster J R P, Jones R A L 2011 Langmuir 27 11000
 Google Scholar
Google Scholar
[21] Dunderdale G J, Fairclough J 2013 Langmuir 29 3628
 Google Scholar
Google Scholar
[22] Lee S J, Park K 1994 J. Vac. Sci. Technol. A 12 2949
 Google Scholar
Google Scholar
[23] Wittemann A, Ballauff M 2006 Physical Chemistry Chemical Physics 8 5269
[24] Koski J P, Frischknecht A L 2018 ACS Nano. 12 1664
 Google Scholar
Google Scholar
[25] Russo D, Plazanet M, Teixeira J, Moulin M, Härtlein M, Wurm F R, Steinbach T 2016 Biomacromolecules 17 141
 Google Scholar
Google Scholar
[26] Feng C, Liu Y, Ren C L 2018 Soft Matter 14 6521
 Google Scholar
Google Scholar
[27] Hui O, Xia Z, Jiang Z 2009 Nanotechnology 20 195703
 Google Scholar
Google Scholar
[28] Tong C H 2015 J. Chem. Phys. 143 79
[29] Ding H D, Duan C, Tong C H 2017 J. Chem. Phys. 146 034901
 Google Scholar
Google Scholar
[30] Baker S L, Murata H, Kaupbayeva B, Tasbolat A, Russell A J 2019 Biomacromolecules 20 2392
 Google Scholar
Google Scholar
[31] Boubeta F M, Soler-Illia G, Tagliazucchi M 2018 Langmuir 34 15727
 Google Scholar
Google Scholar
[32] Szleifer I 1997 Phys. A:Statist. Mech. Its Appl. 244 370
 Google Scholar
Google Scholar
[33] Bratek-Skicki A, Eloy P, Morga M, Dupont-Gillain C 2018 Langmuir 34 3037
 Google Scholar
Google Scholar
[34] Gong K, Pan C, He K, Zhu H K, Chen L J, Hou M X, Wang Y M 2019 J. Appl. Polym. Sci. 136 48135
 Google Scholar
Google Scholar
[35] 赵新军, 李九智, 石铭芸, 马超 2019 68 214701
Zhao X J, Li J Z, Shi M Y, M C 2019 Acta Phys. Sin. 68 214701
[36] Ren C L, Ma Y Q 2006 J. Am. Chem. Soc. 128 2733
[37] Mumtaz F, Chen C S, Zhu H K, Pan C, Wang Y M 2018 Appl. Surf. Sci. 439 148
 Google Scholar
Google Scholar
[38] Delcroix M F, Laurent S, Huet G L, Dupont-Gillain C C 2015 Acta Biomater. 11 68
 Google Scholar
Google Scholar
-
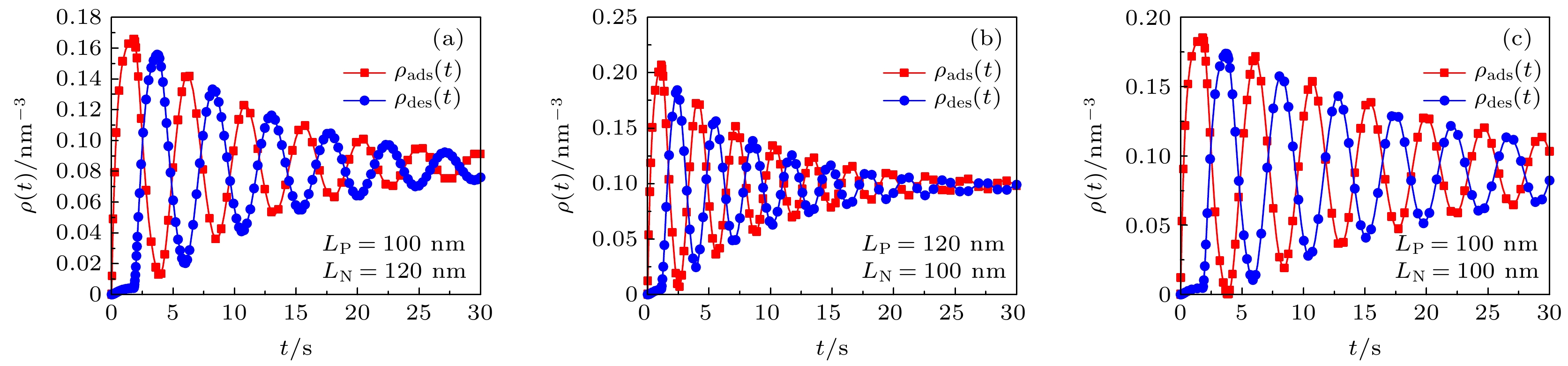
图 4 高分子混合刷中吸附/脱附的蛋白质分子数密度随时间周期性演化关系图. 在子图 (a) 中
$\tau = 1.8\, \, {\rm{s}}$ 、$\tau = 0.3\, \, {\rm{s}}$ ; 在子图 (b)−(e) 中$\tau $ 、$' $ 为变量Figure 4. The number density of adsorbed/desorbed protein molecules in the polymer mixed brush evolves periodically with time. In subfigure (a),
$\tau = 1.8\, \, {\rm{s}}$ 、$\tau = 0.3\, \, {\rm{s}}$ ; in subfigure (b)−(e),$\tau $ and$' $ are variable.图 5 不同
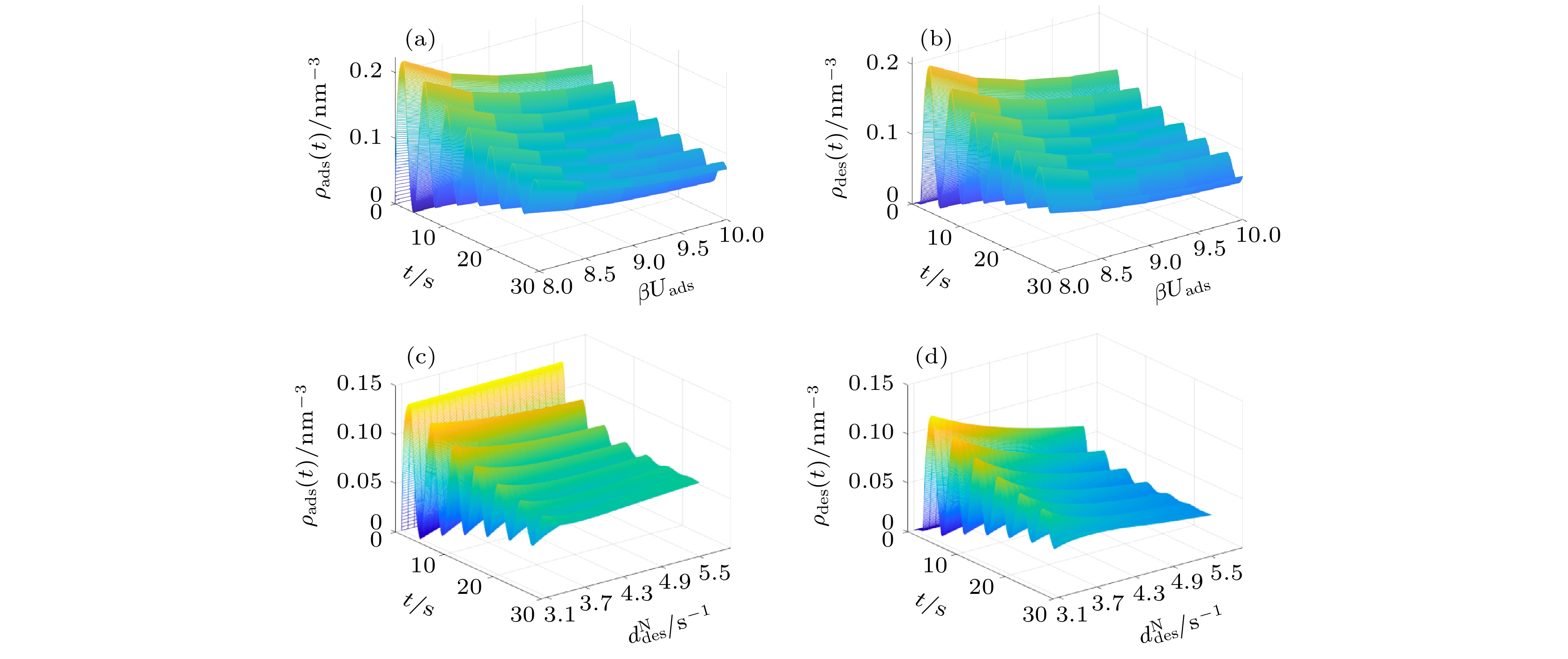
$\beta {U_{{\rm{ads}}}}$ (a)−(b) 与不同$d_{{\rm{des}}}^{\rm{N}}$ (c)−(d) 条件下, 高分子混合刷中吸附/脱附的蛋白质分子数密度随时间演化的动力学Figure 5. The dynamics of the number density of adsorbed/desorbed protein molecules by the polymer mixed brush with time evolution at different
$\beta {U_{{\rm{ads}}}}$ (a)−(b) and different$d_{{\rm{des}}}^{\rm{N}}$ (c)−(d).表 1 不同的部分参数所对应的高分子混合刷对蛋白质吸附/解吸附的状态特性
Table 1. The state characteristics of protein adsorption/desorption by polymer mixed brushes corresponding to some different parameters.
$ {\rho _{{{\rm{pro, bulk}}}}} $ $K_{ { {\rm{des} } } }'/{ { {\rm{s} } }^{ - 1} }$ $ d_{{{\rm{des}}}}^{{\rm{N}}}/{{{\rm{s}}}^{ - 1}} $ $ d_{{{\rm{ads}}}}^{{\rm{P}}}/{{{\rm{s}}}^{ - 1}} $ $ d_{{{\rm{des}}}}^{{\rm{P}}}/{{{\rm{s}}}^{ - 1}} $ $ \tau /{{\rm{s}}} $ $ \tau '/{{\rm{s}}} $ 吸附/脱附特性 $ 0.4 $ $ 0.96\, $ $ 6.0\, $ $ 2.0\, $ $ 3.0\, $ $ 0.3\, $ $ 0.3\, $ 部分吸附/脱附 $ 0.4 $ $ 2.0\, $ $ 0.1 $ $ 27.5\, $ $ 0.01\, $ $ 0.02\, $ $ 0.3\, $ 完全吸附/脱附 $ 0.7 $ $ 2.96\, $ $ 3.1\, $ $ 0.68 $ $ 0.08 $ $ 1.8\, $ $ 0.3\, $ 周期性吸附/脱附 -
[1] Tan J S, Martic P A 1990 J. Colloid Interf. Sci. 136 415
 Google Scholar
Google Scholar
[2] Steadman B L, Thompson K C, Middaugh C R, Matsuno K, Vrona S, Lawson E Q, Lewis R V 1992 Biotechn. Bioengin. 40 8
 Google Scholar
Google Scholar
[3] Norde W, Gage D 2004 Langmuir 20 4162
 Google Scholar
Google Scholar
[4] Moerz S T, Huber P 2015 J. Phys. Chem. C 119 27072
 Google Scholar
Google Scholar
[5] Ladam G, Gergely C, Senger B, Decher G, Voegel J C, Schaaf P, Cuisinier F J 2000 Biomacromolecules 1 674
 Google Scholar
Google Scholar
[6] Wittemann A, Ballauff M 2006 Phys. Chem. Chem. Phys. 8 5269
 Google Scholar
Google Scholar
[7] Yang J, Hua Z, Wang T, Wu B, Liu G, Zhang G Z 2015 Langmuir 31 6078
 Google Scholar
Google Scholar
[8] Delcroix M F, Huet G L, Conard T, Demoustier-Champagne S, Prez F E D, Landoulsi J, Dupont-Gillain C C 2013 Biomacromolecules 14 215
 Google Scholar
Google Scholar
[9] Bratek-Skicki A, Cristaudo V, Savocco J, et al. 2019 Biomacromolecules 20 778
 Google Scholar
Google Scholar
[10] Pan C, Liu X R, Gong K, Mumtaz F, Wang Y M 2018 J. Mater. Chem. B 6 556
[11] Halperin A 1999 Langmuir 15 2525
 Google Scholar
Google Scholar
[12] Fang F, Satulovsky J, Szleifer I 2005 Biophys. J. 89 1516
 Google Scholar
Google Scholar
[13] Su X H, Lei Q L, Ren C L 2015 Chin. Phys. B 24 113601
 Google Scholar
Google Scholar
[14] Biesheuvel P M, Wittemann A 2005 J. Phys. Chem. B 109 4209
[15] Biesheuvel P M, Leermakers F, Stuart M 2006 Phys. Rev. E 73 011802
[16] Xu Y L, Chen X Q, Chen H Y, Xu S H, Liu H L, Hu Y 2012 Mol. Simulat. 38 274
 Google Scholar
Google Scholar
[17] Szleifer I, Carignano M A 2000 Macromol. Rapid Commun. 21 423
 Google Scholar
Google Scholar
[18] Fang F, Szleifer I 2001 Biophys. J. 80 2568
 Google Scholar
Google Scholar
[19] Fang F, Szleifer I 2002 Langmuir 18 5497
 Google Scholar
Google Scholar
[20] Weir M P, Heriot S Y, Martin S J, Parnell A J, Holt S A, Webster J R P, Jones R A L 2011 Langmuir 27 11000
 Google Scholar
Google Scholar
[21] Dunderdale G J, Fairclough J 2013 Langmuir 29 3628
 Google Scholar
Google Scholar
[22] Lee S J, Park K 1994 J. Vac. Sci. Technol. A 12 2949
 Google Scholar
Google Scholar
[23] Wittemann A, Ballauff M 2006 Physical Chemistry Chemical Physics 8 5269
[24] Koski J P, Frischknecht A L 2018 ACS Nano. 12 1664
 Google Scholar
Google Scholar
[25] Russo D, Plazanet M, Teixeira J, Moulin M, Härtlein M, Wurm F R, Steinbach T 2016 Biomacromolecules 17 141
 Google Scholar
Google Scholar
[26] Feng C, Liu Y, Ren C L 2018 Soft Matter 14 6521
 Google Scholar
Google Scholar
[27] Hui O, Xia Z, Jiang Z 2009 Nanotechnology 20 195703
 Google Scholar
Google Scholar
[28] Tong C H 2015 J. Chem. Phys. 143 79
[29] Ding H D, Duan C, Tong C H 2017 J. Chem. Phys. 146 034901
 Google Scholar
Google Scholar
[30] Baker S L, Murata H, Kaupbayeva B, Tasbolat A, Russell A J 2019 Biomacromolecules 20 2392
 Google Scholar
Google Scholar
[31] Boubeta F M, Soler-Illia G, Tagliazucchi M 2018 Langmuir 34 15727
 Google Scholar
Google Scholar
[32] Szleifer I 1997 Phys. A:Statist. Mech. Its Appl. 244 370
 Google Scholar
Google Scholar
[33] Bratek-Skicki A, Eloy P, Morga M, Dupont-Gillain C 2018 Langmuir 34 3037
 Google Scholar
Google Scholar
[34] Gong K, Pan C, He K, Zhu H K, Chen L J, Hou M X, Wang Y M 2019 J. Appl. Polym. Sci. 136 48135
 Google Scholar
Google Scholar
[35] 赵新军, 李九智, 石铭芸, 马超 2019 68 214701
Zhao X J, Li J Z, Shi M Y, M C 2019 Acta Phys. Sin. 68 214701
[36] Ren C L, Ma Y Q 2006 J. Am. Chem. Soc. 128 2733
[37] Mumtaz F, Chen C S, Zhu H K, Pan C, Wang Y M 2018 Appl. Surf. Sci. 439 148
 Google Scholar
Google Scholar
[38] Delcroix M F, Laurent S, Huet G L, Dupont-Gillain C C 2015 Acta Biomater. 11 68
 Google Scholar
Google Scholar
Catalog
Metrics
- Abstract views: 6154
- PDF Downloads: 95
- Cited By: 0














 DownLoad:
DownLoad: