-
Homologous recombination (HR) which is important for maintaining the genetic stability of the cell is mainly mediated by recombinase. As a critical recombinase exists in prokaryotic organism, RecA has been widely studied. RecA could bind single strand DNA to form a filament structure to perform strand invasion and exchange with homologous DNA. The basic strand exchange step is a pathway to investigating the mechanism of strand exchange process. However, the specific step length is still controversial. The mainstream view is to regard the basic step as 3 bp, which sounds reasonable because the basic unit of RecA filament is a RecA monomer which binds 3 base pairs, but our previous study found that the most probable basic strand exchange step is 9 bp. In this research, we set up a series of experiments to demonstrate that the basic strand exchange step is 9 bp and how the mismatch base pairs in 9 bp affect the strand exchange process. We ues digestion protection to confirm whether the strand exchange process is completed and we employ a magnetic tweezer to measure how many base pairs are exchanged by strand exchange process. The digestion protection experiments show the strand exchange cannot overcome 33% mismatch sequence, but according to the result that strand exchange could overcome a single mismatch base pair, so that the step is not 3 bp. According to strand exchange length of 33% mismatch sequence, we find a 9 bp interval between the main peaks, which implies that the strand exchange step should be 9 bp rather than 3 bp. We also use continuous mismatch base pairs instead of periodic mismatch sequence to see whether the strand exchange process can be overcome. We find that the more the mismatch base pairs, the harder the strand exchange process passes across. Homology degree and its distribution both affect the strand exchange process. In general, the results suggest that the strand exchange step mediated by RecA is 9 bp, and the number of mismatched base pairs and their distribution influence the strand exchange process. The combination of digestion protection assay and magnetic tweezers can further take advantage of single molecule techniques to investigate mechanism of HR.
-
Keywords:
- RecA /
- single-molecule biophysics /
- homologous recombination /
- mismatch base pair
[1] Blaikley E J, Tinline-Purvis H, Kasparek T R, Marguerat S, Sarkar S, Hulme L, Hussey S, Wee B Y, Deegan R S, Walker C A, Pai C C, Bahler J, Nakagawa T, Humphrey T C 2014 Nucleic Acids Res. 42 5644
 Google Scholar
Google Scholar
[2] Sakofsky C J, Malkova A 2017 Crit. Rev. Biochem. Mol. Biol. 52 395
 Google Scholar
Google Scholar
[3] Chen Z, Yang H, Pavletich N P 2008 Nature 453 489
 Google Scholar
Google Scholar
[4] Kim S H 2018 Methods Enzymol. 600 233
 Google Scholar
Google Scholar
[5] Lu C H, Li H W 2017 ChemPhysChem 18 584
 Google Scholar
Google Scholar
[6] Danilowicz C, Hermans L, Coljee V, Prevost C, Prentiss M 2017 Nucleic Acids Res. 45 8448
 Google Scholar
Google Scholar
[7] Ragunathan K, Joo C, Ha T 2011 Structure 19 1064
 Google Scholar
Google Scholar
[8] Lee J Y, Terakawa T, Qi Z, Steinfeld J B, Redding S, Kwon Y, Gaines W A, Zhao W, Sung P, Greene E C 2015 Science 349 977
 Google Scholar
Google Scholar
[9] Peacock V A, Yang D, Danilowicz C, Feinstein E, Pollock N, McShan S, Coljee V, Prentiss M 2012 Nucleic Acids Res. 40 10441
 Google Scholar
Google Scholar
[10] Danilowicz C, Feinstein E, Conover A, Coljee V W, Vlassakis J, Chan Y L, Bishop D K, Prentiss M 2012 Nucleic Acids Res. 40 1717
 Google Scholar
Google Scholar
[11] Jiang L L, Prentiss M 2014 Phys. Rev. E 90 022704
[12] Prentiss M, Prevost C, Danilowicz C 2015 Crit. Rev. Biochem. Mol. Biol. 50 453
 Google Scholar
Google Scholar
[13] 张宇微, 颜燕, 农大官, 徐春华, 李明 2016 65 218702
 Google Scholar
Google Scholar
Zhang Y W, Yan Y, Nong D G, Xu C H, Li M 2016 Acta Phys. Sin. 65 218702
 Google Scholar
Google Scholar
[14] Danilowicz C B, Coljee V, Bouzig C, Conroy R S, Lubensky D, Sarkar A, Nelson D R, Prentiss M 2003 Biophys. J. 84 301a
[15] Winkleman A, Gudiksen K L, Ryan D, Whitesides G M, Greenfield D, Prentiss M 2004 Appl. Phys. Lett. 85 2411
 Google Scholar
Google Scholar
[16] Lipfert J, Hao X M, Dekker N H 2009 Biophys. J. 96 5040
 Google Scholar
Google Scholar
[17] 马建兵, 翟永亮, 农大官, 李菁华, 付航, 张兴华, 李明, 陆颖, 徐春华 2018 67 148702
 Google Scholar
Google Scholar
Ma J B, Zhai Y L, Nong D G, Li J H, Fu H, Zhang X H, Li M, Lu Y, Xu C H 2018 Acta Phys. Sin. 67 148702
 Google Scholar
Google Scholar
[18] Le S M, Yao M X, Chen J, Efremov A K, Azimi S, Yan J 2015 Nucleic Acids Res. 43 e113
 Google Scholar
Google Scholar
[19] Wang Y Z, Hou X M, Ju H P, Xiao X, Xi X G, Dou S X, Wang P Y, Li W 2018 Chin. Phys. B 27 068701
 Google Scholar
Google Scholar
[20] 赵振业, 徐春华, 李菁华, 黄星榞, 马建兵, 陆颖 2017 66 188701
 Google Scholar
Google Scholar
Zhao Z Y, Xu C H, Li J H, Huang X Y, Ma J B, Lu Y 2017 Acta Phys. Sin. 66 188701
 Google Scholar
Google Scholar
[21] Lee J Y, Steinfeld J B, Qi Z, Kwon Y, Sung P, Greene E C 2017 J. Biol. Chem. 292 11125
 Google Scholar
Google Scholar
[22] Qi Z, Redding S, Lee J Y, Gibb B, Kwon Y, Niu H, Gaines W A, Sung P, Greene E C 2015 Cell 160 856
 Google Scholar
Google Scholar
[23] Ristic D, Kanaar R, Wyman C 2011 Nucleic Acids Res. 39 155
 Google Scholar
Google Scholar
[24] Smith S B, Cui Y, Bustamante C 1996 Science 271 795
 Google Scholar
Google Scholar
[25] Bustamante C, Marko J F, Siggia E D, Smith S 1994 Science 265 1599
 Google Scholar
Google Scholar
[26] Vlassakis J, Feinstein E, Yang D, Tilloy A, Weiller D, Kates H J, Coljee V, Prentiss M 2013 Phys. Rev. E 87 012702
-
图 1 全匹配酶切保护实验的示意图及酶切结果 (a) 包含错配序列的酶切保护实验示意图, 蓝色部分代表同源序列, 红色部分代表错配序列; (b) 在全匹配序列的情况下酶切保护实验示意图; (c) 酶切实验结果, 分别为全匹配序列和33%错配序列的酶切情况(误差线代表统计误差), 酶切时间为30 min
Figure 1. The enzyme protection assay: (a) The diagram of the enzyme protection assay with mismatch sequence, blue strands represent homologous sequence and red strand represents mismatched sequence; (b) same as (a) when using completely homologous sequence; (c) the results of enzyme protection assay.
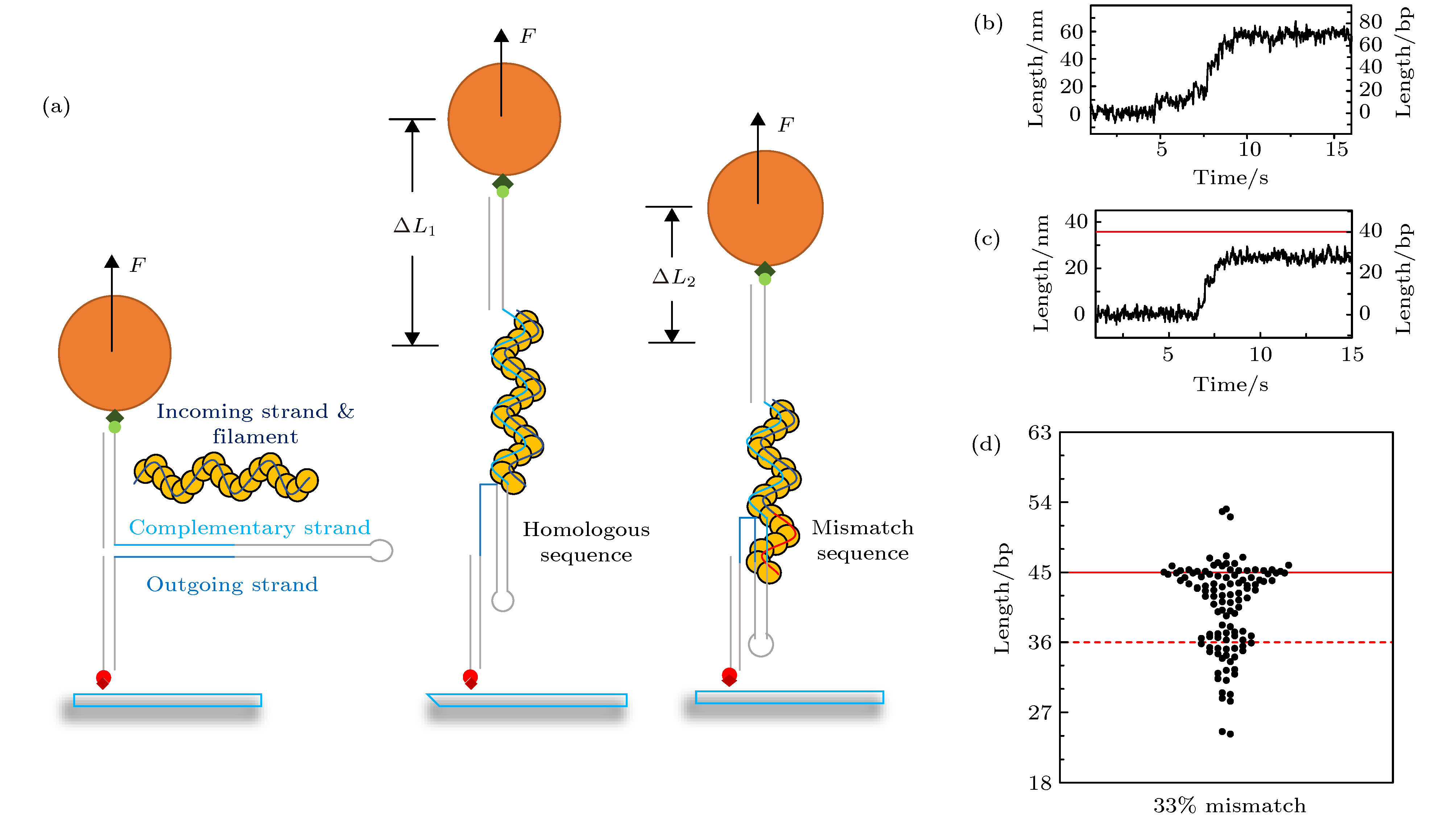
图 2 用磁镊定量测量不同匹配程度序列下的换链长度 (a) 磁镊实验的示意图, 红色链代表错配序列, 蓝色链代表同源序列, 从左到右分别对应链交换反应发生之前, 全匹配序列的链交换反应, 和链交换被错配序列阻挡三种现象; (b) 全匹配序列下的换链曲线, 换链可以进行到末尾; (c) 后24个碱基为部分错配时的换链曲线, 红色为界面位置; (d) 33%错配时的换链长度, 红色实线代表界面位置, 红色虚线为界面前9 bp的位置
Figure 2. Using magnetic tweezers to measure the strand exchange length under different periodic mismatch sequence: (a) From left to right are diagrams of experiments when strand exchange is not happened, strand exchange process under homologous sequence and periodic mismatch sequence, respectively. Red strands represent mismatch sequence and blue strands represent homologous sequence; (b) trace of fully homologous sequence; (c) trace of periodic mismatch sequence; (d) strand exchange length distributions of 33% periodic mismatch sequence, red line marks the interface and red-dashed line marks the position which is 9 bp before the interface.
图 3 使用磁镊方法探究换链是否能够跨过连续错配碱基 (a) 磁镊实验的示意图, 红色线段位置为错配碱基序列, 蓝色线段为同源碱基序列, 从左到右分别是链交换反应之前, 链交换被错配碱基阻碍和链交换跨过错配序列三种现象; (b) 不同连续错配碱基数目时, 越过错配碱基事件数量的百分比(误差线代表统计误差); (c) 33%错配序列和连续3 bp错配序列的碱基位置分布, N为错配碱基数目, 红色代表错配碱基, 蓝色代表同源碱基; (d)同(c)一样, 用来表示N = 2, 4和5时的错配碱基分布
Figure 3. Using magnetic tweezers to investigate if strand exchange could stride across continuous mismatch base pairs: (a) From left to right are diagrams of experiments when strand exchange is not happened, strand exchange process blocked by continuous mismatch base pairs and strand exchange process stride across continuous mismatch base pairs, respectively. Red segments represent mismatch base pairs and blue segments represent homologous base pairs; (b) across probability when facing different continuous sequence; (c) the mismatch base pairs distribution of 33% periodic mismatch sequence and 3 bp continuous mismatch sequence, N represents the number of mismatch base pairs; (d) same as (c) when N = 2, 4 and 5.
-
[1] Blaikley E J, Tinline-Purvis H, Kasparek T R, Marguerat S, Sarkar S, Hulme L, Hussey S, Wee B Y, Deegan R S, Walker C A, Pai C C, Bahler J, Nakagawa T, Humphrey T C 2014 Nucleic Acids Res. 42 5644
 Google Scholar
Google Scholar
[2] Sakofsky C J, Malkova A 2017 Crit. Rev. Biochem. Mol. Biol. 52 395
 Google Scholar
Google Scholar
[3] Chen Z, Yang H, Pavletich N P 2008 Nature 453 489
 Google Scholar
Google Scholar
[4] Kim S H 2018 Methods Enzymol. 600 233
 Google Scholar
Google Scholar
[5] Lu C H, Li H W 2017 ChemPhysChem 18 584
 Google Scholar
Google Scholar
[6] Danilowicz C, Hermans L, Coljee V, Prevost C, Prentiss M 2017 Nucleic Acids Res. 45 8448
 Google Scholar
Google Scholar
[7] Ragunathan K, Joo C, Ha T 2011 Structure 19 1064
 Google Scholar
Google Scholar
[8] Lee J Y, Terakawa T, Qi Z, Steinfeld J B, Redding S, Kwon Y, Gaines W A, Zhao W, Sung P, Greene E C 2015 Science 349 977
 Google Scholar
Google Scholar
[9] Peacock V A, Yang D, Danilowicz C, Feinstein E, Pollock N, McShan S, Coljee V, Prentiss M 2012 Nucleic Acids Res. 40 10441
 Google Scholar
Google Scholar
[10] Danilowicz C, Feinstein E, Conover A, Coljee V W, Vlassakis J, Chan Y L, Bishop D K, Prentiss M 2012 Nucleic Acids Res. 40 1717
 Google Scholar
Google Scholar
[11] Jiang L L, Prentiss M 2014 Phys. Rev. E 90 022704
[12] Prentiss M, Prevost C, Danilowicz C 2015 Crit. Rev. Biochem. Mol. Biol. 50 453
 Google Scholar
Google Scholar
[13] 张宇微, 颜燕, 农大官, 徐春华, 李明 2016 65 218702
 Google Scholar
Google Scholar
Zhang Y W, Yan Y, Nong D G, Xu C H, Li M 2016 Acta Phys. Sin. 65 218702
 Google Scholar
Google Scholar
[14] Danilowicz C B, Coljee V, Bouzig C, Conroy R S, Lubensky D, Sarkar A, Nelson D R, Prentiss M 2003 Biophys. J. 84 301a
[15] Winkleman A, Gudiksen K L, Ryan D, Whitesides G M, Greenfield D, Prentiss M 2004 Appl. Phys. Lett. 85 2411
 Google Scholar
Google Scholar
[16] Lipfert J, Hao X M, Dekker N H 2009 Biophys. J. 96 5040
 Google Scholar
Google Scholar
[17] 马建兵, 翟永亮, 农大官, 李菁华, 付航, 张兴华, 李明, 陆颖, 徐春华 2018 67 148702
 Google Scholar
Google Scholar
Ma J B, Zhai Y L, Nong D G, Li J H, Fu H, Zhang X H, Li M, Lu Y, Xu C H 2018 Acta Phys. Sin. 67 148702
 Google Scholar
Google Scholar
[18] Le S M, Yao M X, Chen J, Efremov A K, Azimi S, Yan J 2015 Nucleic Acids Res. 43 e113
 Google Scholar
Google Scholar
[19] Wang Y Z, Hou X M, Ju H P, Xiao X, Xi X G, Dou S X, Wang P Y, Li W 2018 Chin. Phys. B 27 068701
 Google Scholar
Google Scholar
[20] 赵振业, 徐春华, 李菁华, 黄星榞, 马建兵, 陆颖 2017 66 188701
 Google Scholar
Google Scholar
Zhao Z Y, Xu C H, Li J H, Huang X Y, Ma J B, Lu Y 2017 Acta Phys. Sin. 66 188701
 Google Scholar
Google Scholar
[21] Lee J Y, Steinfeld J B, Qi Z, Kwon Y, Sung P, Greene E C 2017 J. Biol. Chem. 292 11125
 Google Scholar
Google Scholar
[22] Qi Z, Redding S, Lee J Y, Gibb B, Kwon Y, Niu H, Gaines W A, Sung P, Greene E C 2015 Cell 160 856
 Google Scholar
Google Scholar
[23] Ristic D, Kanaar R, Wyman C 2011 Nucleic Acids Res. 39 155
 Google Scholar
Google Scholar
[24] Smith S B, Cui Y, Bustamante C 1996 Science 271 795
 Google Scholar
Google Scholar
[25] Bustamante C, Marko J F, Siggia E D, Smith S 1994 Science 265 1599
 Google Scholar
Google Scholar
[26] Vlassakis J, Feinstein E, Yang D, Tilloy A, Weiller D, Kates H J, Coljee V, Prentiss M 2013 Phys. Rev. E 87 012702
Catalog
Metrics
- Abstract views: 10828
- PDF Downloads: 110
- Cited By: 0















 DownLoad:
DownLoad: