-
不同结构类型的蛋白质的力学稳定性和去折叠动力学有显著的差异, 其中全部由α螺旋构成的蛋白质通常在较小的拉力作用下就会发生快速去折叠, 需要能够精准控制皮牛量级拉力的实验手段来进行定量研究. 酰基辅酶A结合蛋白(ACBP)是一种研究全α螺旋结构蛋白折叠/去折叠动力学性质的模型蛋白, 其由86个氨基酸残基形成一种由4个α螺旋组成的螺旋束结构. 本文利用磁镊对ACBP进行了恒定力加载速率的拉伸实验, 得到了不同力加载速率下的去折叠力的分布. 不同数据分析方法都显示ACBP具有超长的去折叠距离. 对比分子动力学模拟拉伸的结果, ACBP的过渡态中整个N端α螺旋和部分的C端α螺旋发生了去折叠转变. 本研究显示单分子拉伸实验与分子动力学模拟相结合是揭示蛋白质构象转变的分子机制的可靠研究方法.There are significant differences in mechanical stability and unfolding dynamics among proteins with different structural compositions. Compared with proteins with β-sheets and subjected to shearing forces, proteins that are composed entirely of α-helices often undergo rapid unfolding under low stretching forces, thus requiring quantitative studies by using experimental tools that can precisely control forces on a pico-Newton scale. Magnetic tweezers with intrinsic force-control capability and great stability for long-time continuous measurement are suitable to measure force-induced conformation transitions of protein subjected to low forces of several pico-Newton. Acyl-CoA binding protein (ACBP) is a model protein used to study the folding/unfolding kinetics of complete α-helices protein. It is composed of 86 amino acid residues, forming a helical bundle of four α-helices. When its N- and C-terminal are stretched, the first and last α-helix are subjected to shear force in parallel. Previous biochemical studies showed that ACBP folding and unfolding in a two-state manner. In this paper, we use magnetic tweezers to stretch ACBP from its N- and C-end and obtain the distribution of the unfolding force at different loading rates ranging from 0.25 pN/s to 4 pN/s. The most probable unfolding forces are all less than 10 pN, which confirms that ACBP is not mechanically stable. At a constant loading rate, the unfolding force distribution and the most probable unfolding force as a function of loading rate have well-defined analytical formulas based on Bell’s model. Therefore, the experimental results of unfolding force can be fitted directly to obtain the important kinetic parameter of unfolding distance which is defined as the difference in extension between the native state and the transition state. Data analysis shows that ACBP has an extraordinarily long unfolding distance of 7.8 nm. Steered molecular dynamics simulations of ACBP stretching gives the transition state with N-terminal α-helix fully unfolded and C-terminal α-helix partially unfolded, which is consistent with the long unfolding distance obtained in the experiment on magnetic tweezers. According to the simulation results, the unfolding of α-helices is less cooperative than that of β-sheet structures. This characteristic makes α-helix proteins sensitive to mechanical forces, rendering them suitable as force sensors in cells. This study shows that single-molecule stretching experiment combined with molecular dynamics simulations is a reliable method to reveal the molecular mechanism of protein conformationtransitions under stretching forces.
[1] Ayoub R, Lee Y 2021 Proteins Struct. Funct. Bioinf. 89 648
 Google Scholar
Google Scholar
[2] Lane T J 2023 Nat. Methods 20 170
 Google Scholar
Google Scholar
[3] Kaiser C M, Goldman D H, Chodera J D, Tinoco I, Bustamante C 2011 Science 334 1723
 Google Scholar
Google Scholar
[4] Aubin-Tam M E, Olivares A O, Sauer R T, Baker T A, Lang M J 2011 Cell 145 257
 Google Scholar
Google Scholar
[5] Uddin M S, Al Mamun A, Rahman M A, Behl T, Perveen A, Hafeez A, Bin-Jumah M N, Abdel-Daim M M, Ashraf G M 2020 Curr. Top. Med. Chem. 20 2380
 Google Scholar
Google Scholar
[6] Tan J M M, Wong E S P, Lim K L 2009 Antioxid. Redox Signaling. 11 2119
 Google Scholar
Google Scholar
[7] Kumar T K S, Sivaraman T, Samuel D, Srisailam S, Ganesh G, Hsieh H C, Hung K W, Peng H J, Ho M C, Arunkumar A I, Yu C 2000 J. Chin. Chem. Soc. 47 1009
 Google Scholar
Google Scholar
[8] Fung A, Li P, Godoy-Ruiz R, Sanchez-Ruiz J M, Muñoz V 2008 J. Am. Chem. Soc. 130 7489
 Google Scholar
Google Scholar
[9] Dill K A, MacCallum J L 2012 Science 338 1042
 Google Scholar
Google Scholar
[10] Ma X, Sun H, Hong H, Guo Z, Su H, Chen H 2022 Phys. Rev. E 106 024404
 Google Scholar
Google Scholar
[11] Jagannathan B, Elms P J, Bustamante C, Marqusee S 2012 Proc. Natl. Acad. Sci. 109 17820
 Google Scholar
Google Scholar
[12] Pierse C A, Dudko O K 2017 Phys. Rev. Lett. 118 088101
 Google Scholar
Google Scholar
[13] Piras M, Gerosa C, Fanni D, Cau F, Coni P, Murru R, Denotti G, Orrù G, Scano A, Ledda F, Van Eyken P, Coghe F, Faa G, Castagnola M 2022 Eur. Rev. Med. Pharmacol. Sci. 26 3301
 Google Scholar
Google Scholar
[14] Poulsen F M, Kragelund B B, Osmark P, Neergaard T B, Schiødt J, Kristiansen K, Knudsen J 1999 Nat. Struct. Biol. 6 594
 Google Scholar
Google Scholar
[15] Heidarsson P O, Valpapuram I, Camilloni C, Imparato A, Tiana G, Poulsen F M, Kragelund B B, Cecconi C 2012 J. Am. Chem. Soc. 134 17068
 Google Scholar
Google Scholar
[16] Mandal S S 2020 ACS Omega 5 11271
 Google Scholar
Google Scholar
[17] Neuman K C, Nagy A 2008 Nat. Methods 5 491
 Google Scholar
Google Scholar
[18] Qian H, Chen H, Yan J 2016 Acta Phys. Sin. 65 188706
 Google Scholar
Google Scholar
[19] Chen H, Fu H, Zhu X, Cong P, Nakamura F, Yan J 2011 Biophys. J. 100 517
 Google Scholar
Google Scholar
[20] Miao Y, Feixas F, Eun C, McCammon J A 2016 J. Comput. Chem. 37 1536
 Google Scholar
Google Scholar
[21] Cao Y, Lam C, Wang M, Li H 2006 Angew. Chem. Int. Ed. 45 642
 Google Scholar
Google Scholar
[22] Guo Z, Hong H, Yuan G, Qian H, Li B, Cao Y, Wang W, Wu C X, Chen H 2020 Phys. Rev. Lett. 125 198101
 Google Scholar
Google Scholar
[23] Hong H, Guo Z, Sun H, Yu P, Su H, Ma X, Chen H 2021 Commun. Chem. 4 156
 Google Scholar
Google Scholar
[24] Zhang X, Guo Z, Yu P, Li Q, Zhou X, Chen H 2020 Chin. Phys. B 29 078701
 Google Scholar
Google Scholar
[25] Chen H, Yuan G, Winardhi R S, Yao M, Popa I, Fernandez J M, Yan J 2015 J. Am. Chem. Soc. 137 3540
 Google Scholar
Google Scholar
[26] Yuan G, Le S, Yao M, Qian H, Zhou X, Yan J, Chen H 2017 Angew. Chem. Int. Ed. 56 5490
 Google Scholar
Google Scholar
[27] Cao Y, Li H 2007 Nat. Mater. 6 109
 Google Scholar
Google Scholar
[28] Lu H, Schulten K 2000 Biophys. J. 79 51
 Google Scholar
Google Scholar
[29] Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E 1997 Science 276 1109
 Google Scholar
Google Scholar
[30] Yao M, Goult B T, Chen H, Cong P, Sheetz M P, Yan J 2014 Sci. Rep. 4 4610
 Google Scholar
Google Scholar
[31] Yao M, Goult B T, Sheetz M P, Yan J 2017 Biophys. J. 112 267a
 Google Scholar
Google Scholar
[32] Yao M, Qiu W, Liu R, Efremov A K, Cong P, Seddiki R, Payre M, Lim C T, Ladoux B, Mège R M, Yan J 2014 Nat. Commun. 5 4525
 Google Scholar
Google Scholar
-
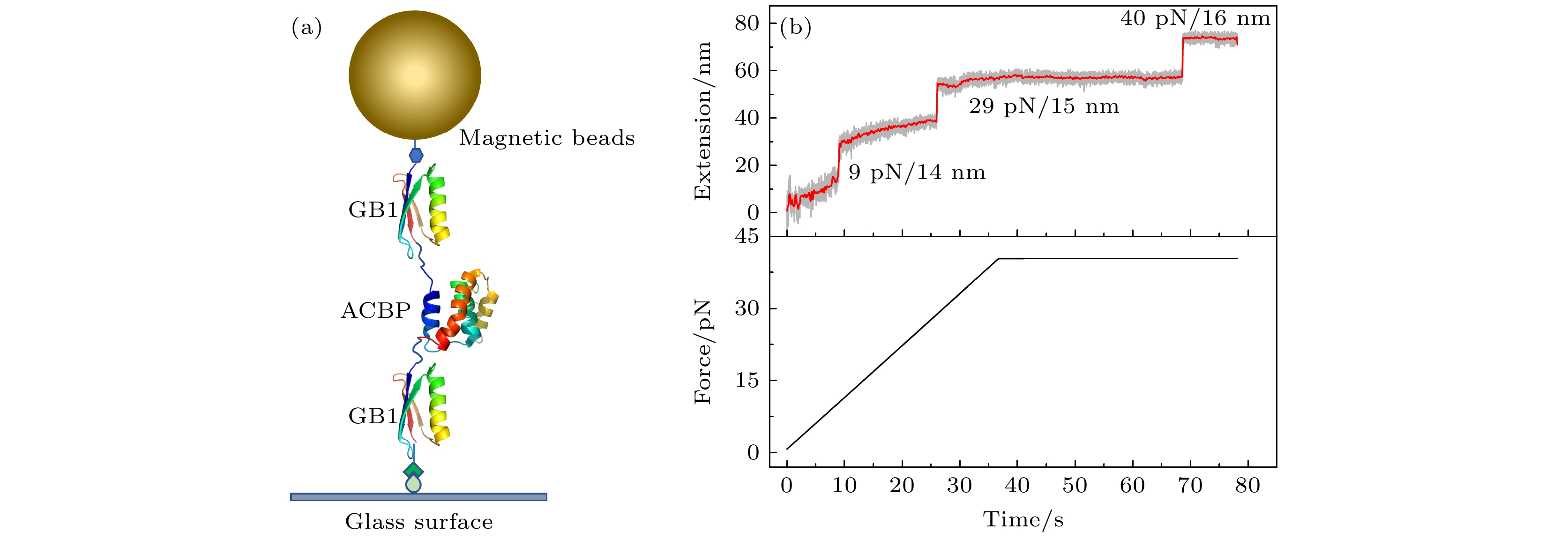
图 1 单分子磁镊实验的结构示意图及典型的拉力-伸长曲线 (a)磁镊拉伸蛋白的结构示意图, 目的蛋白ACBP (PDB ID:2ABD)两端分别连接一个GB1蛋白作为特征信号, 通过顶部的磁球施加竖直向上的拉力; (b) ACBP蛋白构建体的典型的拉力-伸长曲线, 拉力以1 pN/s的恒定加载速率从0.5 pN增加到40 pN并保持在40 pN, 通过伸长的测量能够依次观察到ACBP和2个GB1的去折叠信号
Fig. 1. Schematic figure of a single molecule magnetic tweezers experiment and typical force-extension curve. (a) Schematic figure of magnetic tweezers setup to stretch the designed ACBP protein construct. ACBP (PDB ID:2ABD) is sandwiched between two GB1 proteins which provide fingerprint signals to identify correct protein tether. Vertical upward force is applied through the top magnetic bead. (b) Typical force-extension curve of ACBP protein construct. Force increased from 0.5 pN to 40 pN with a constant loading rate of 1 pN/s, and kept at 40 pN. The extension time course shows the unfolding signals of ACBP and GB1 protein domains sequentially.
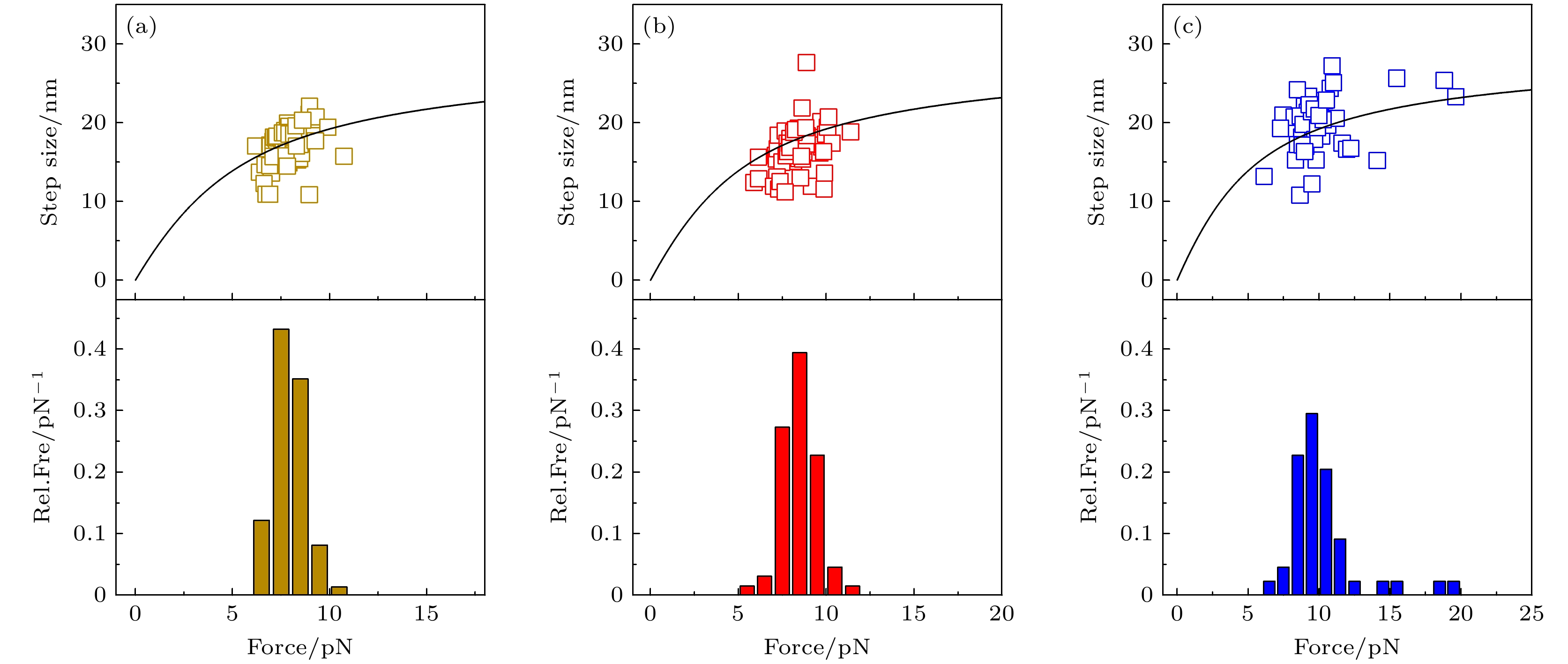
图 2 加载速率分别为0.25 pN/s (a), 1 pN/s (b)和4 pN/s (c)下, 去折叠步长和去折叠力的统计结果. 上面板为去折叠步长与拉力的散点图, 黑色实线为根据多肽链的虫链模型得到的去折叠步长的理论曲线; 下面板为去折叠力分布的相对频率(relative frequency, Rel.Fre).
Fig. 2. Results of step size and unfolding force at loading rates of 0.25 pN/s (a), 1 pN/s (b) and 4 pN/s (c). Upper panel shows scattered data points of the step size and corresponding unfolding force. Solid black line shows theoretical curve obtained from worm-like chain model of polypeptide. Lower panel shows relative frequency (Rel.Fre) of unfolding forces.
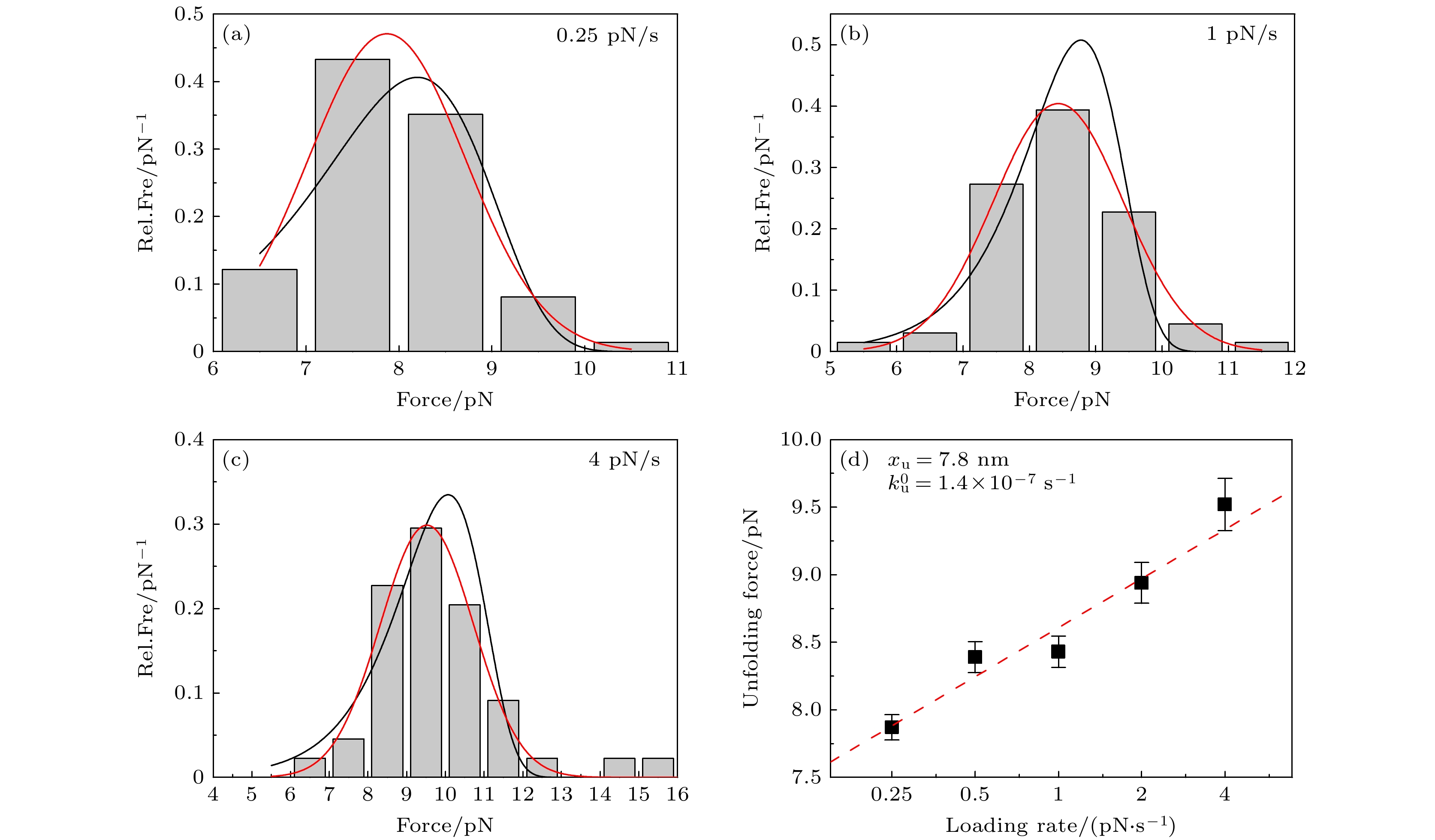
图 3 不同加载速率下的ACBP去折叠力的分析 (a)—(c) 0.25, 1, 4 pN/s的拉力加载速率下, ACBP去折叠力的分布直方图, 红线和黑线分别对应高斯函数和(1)式的拟合曲线; (d)高斯函数拟合得到的最可几去折叠力与拉力加载速率的依赖关系, 红色虚线给出(2)式的拟合结果.
Fig. 3. Analysis of ACBP unfolding force at different loading rates: Distribution histograms of ACBP unfolding force for loading rates of 0.25 pN/s (a), 1 pN/s (b) and 4 pN/s (c), respectively. Red and black curves are fitting curve of Gaussian function and Eq. (1), respectively; (d) most probable unfolding force obtained by fitting of Gaussian function is a function of loading rates. Red dashed line shows fitting curve of Eq. (2).
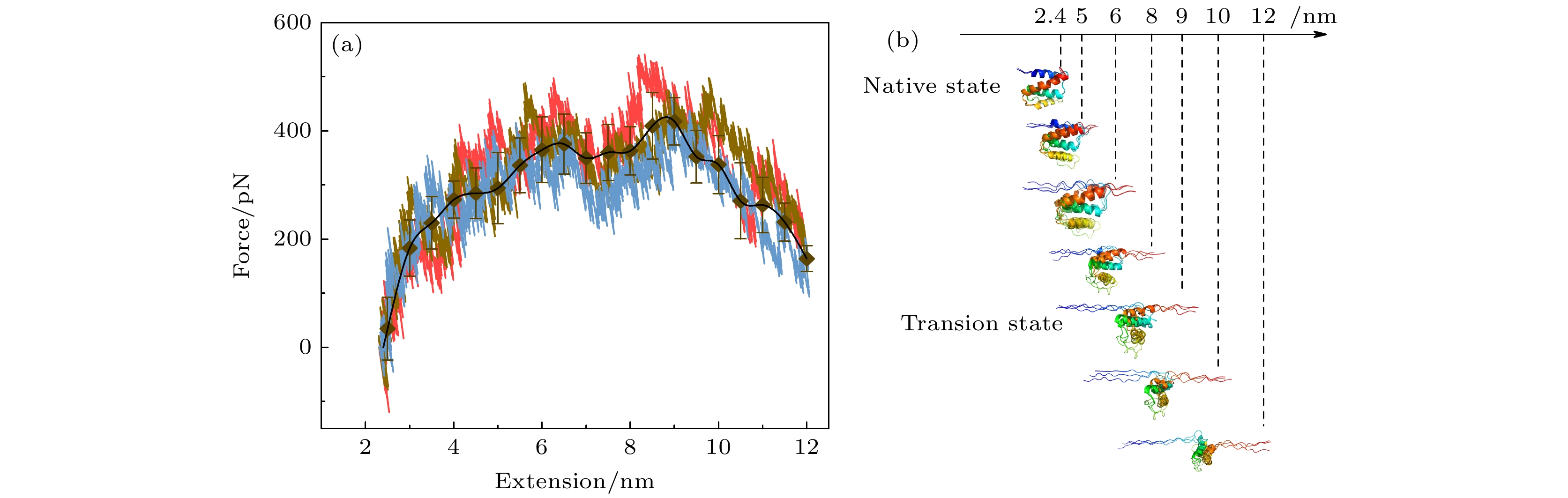
图 4 分子动力学模拟中的拉力-伸长曲线以及蛋白质去折叠过程中的构象变化 (a) 3次独立的分子动力学模拟的拉力伸长曲线用三种颜色给出, 黑色图标是三条曲线的平均值. 在开始阶段, 力随着拉伸的进行逐渐增加, 当N-C距离超过9.0 nm后拉力骤降, 以此判断过渡态构象的伸长约为9.0 nm. (b) ACBP的去折叠过程中的快照图, 去折叠开始于N端, 过渡态构象对应结构的N端α螺旋发生了完全去折叠, C端α螺旋发生了部分去折叠
Fig. 4. Force-extension curves in molecular dynamics simulations and conformational snapshots during unfolding process. (a) Force-extension curves from three independent molecular dynamics simulations are shown in three different colors. Black symbol and black line show the average extension. At the beginning of stretching, force increases gradually. When extension exceeds 9.0 nm, force drops abruptly, which help to identify the transition state with extension of about 9.0 nm. (b) Snapshots during unfolding process of ACBP. Unfolding begins at N-terminal α-helix. Transition state has completely unfolded N-terminal α-helix and partial unfolded C-terminal α-helix.
-
[1] Ayoub R, Lee Y 2021 Proteins Struct. Funct. Bioinf. 89 648
 Google Scholar
Google Scholar
[2] Lane T J 2023 Nat. Methods 20 170
 Google Scholar
Google Scholar
[3] Kaiser C M, Goldman D H, Chodera J D, Tinoco I, Bustamante C 2011 Science 334 1723
 Google Scholar
Google Scholar
[4] Aubin-Tam M E, Olivares A O, Sauer R T, Baker T A, Lang M J 2011 Cell 145 257
 Google Scholar
Google Scholar
[5] Uddin M S, Al Mamun A, Rahman M A, Behl T, Perveen A, Hafeez A, Bin-Jumah M N, Abdel-Daim M M, Ashraf G M 2020 Curr. Top. Med. Chem. 20 2380
 Google Scholar
Google Scholar
[6] Tan J M M, Wong E S P, Lim K L 2009 Antioxid. Redox Signaling. 11 2119
 Google Scholar
Google Scholar
[7] Kumar T K S, Sivaraman T, Samuel D, Srisailam S, Ganesh G, Hsieh H C, Hung K W, Peng H J, Ho M C, Arunkumar A I, Yu C 2000 J. Chin. Chem. Soc. 47 1009
 Google Scholar
Google Scholar
[8] Fung A, Li P, Godoy-Ruiz R, Sanchez-Ruiz J M, Muñoz V 2008 J. Am. Chem. Soc. 130 7489
 Google Scholar
Google Scholar
[9] Dill K A, MacCallum J L 2012 Science 338 1042
 Google Scholar
Google Scholar
[10] Ma X, Sun H, Hong H, Guo Z, Su H, Chen H 2022 Phys. Rev. E 106 024404
 Google Scholar
Google Scholar
[11] Jagannathan B, Elms P J, Bustamante C, Marqusee S 2012 Proc. Natl. Acad. Sci. 109 17820
 Google Scholar
Google Scholar
[12] Pierse C A, Dudko O K 2017 Phys. Rev. Lett. 118 088101
 Google Scholar
Google Scholar
[13] Piras M, Gerosa C, Fanni D, Cau F, Coni P, Murru R, Denotti G, Orrù G, Scano A, Ledda F, Van Eyken P, Coghe F, Faa G, Castagnola M 2022 Eur. Rev. Med. Pharmacol. Sci. 26 3301
 Google Scholar
Google Scholar
[14] Poulsen F M, Kragelund B B, Osmark P, Neergaard T B, Schiødt J, Kristiansen K, Knudsen J 1999 Nat. Struct. Biol. 6 594
 Google Scholar
Google Scholar
[15] Heidarsson P O, Valpapuram I, Camilloni C, Imparato A, Tiana G, Poulsen F M, Kragelund B B, Cecconi C 2012 J. Am. Chem. Soc. 134 17068
 Google Scholar
Google Scholar
[16] Mandal S S 2020 ACS Omega 5 11271
 Google Scholar
Google Scholar
[17] Neuman K C, Nagy A 2008 Nat. Methods 5 491
 Google Scholar
Google Scholar
[18] Qian H, Chen H, Yan J 2016 Acta Phys. Sin. 65 188706
 Google Scholar
Google Scholar
[19] Chen H, Fu H, Zhu X, Cong P, Nakamura F, Yan J 2011 Biophys. J. 100 517
 Google Scholar
Google Scholar
[20] Miao Y, Feixas F, Eun C, McCammon J A 2016 J. Comput. Chem. 37 1536
 Google Scholar
Google Scholar
[21] Cao Y, Lam C, Wang M, Li H 2006 Angew. Chem. Int. Ed. 45 642
 Google Scholar
Google Scholar
[22] Guo Z, Hong H, Yuan G, Qian H, Li B, Cao Y, Wang W, Wu C X, Chen H 2020 Phys. Rev. Lett. 125 198101
 Google Scholar
Google Scholar
[23] Hong H, Guo Z, Sun H, Yu P, Su H, Ma X, Chen H 2021 Commun. Chem. 4 156
 Google Scholar
Google Scholar
[24] Zhang X, Guo Z, Yu P, Li Q, Zhou X, Chen H 2020 Chin. Phys. B 29 078701
 Google Scholar
Google Scholar
[25] Chen H, Yuan G, Winardhi R S, Yao M, Popa I, Fernandez J M, Yan J 2015 J. Am. Chem. Soc. 137 3540
 Google Scholar
Google Scholar
[26] Yuan G, Le S, Yao M, Qian H, Zhou X, Yan J, Chen H 2017 Angew. Chem. Int. Ed. 56 5490
 Google Scholar
Google Scholar
[27] Cao Y, Li H 2007 Nat. Mater. 6 109
 Google Scholar
Google Scholar
[28] Lu H, Schulten K 2000 Biophys. J. 79 51
 Google Scholar
Google Scholar
[29] Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E 1997 Science 276 1109
 Google Scholar
Google Scholar
[30] Yao M, Goult B T, Chen H, Cong P, Sheetz M P, Yan J 2014 Sci. Rep. 4 4610
 Google Scholar
Google Scholar
[31] Yao M, Goult B T, Sheetz M P, Yan J 2017 Biophys. J. 112 267a
 Google Scholar
Google Scholar
[32] Yao M, Qiu W, Liu R, Efremov A K, Cong P, Seddiki R, Payre M, Lim C T, Ladoux B, Mège R M, Yan J 2014 Nat. Commun. 5 4525
 Google Scholar
Google Scholar
计量
- 文章访问数: 8479
- PDF下载量: 131
- 被引次数: 0















 下载:
下载: