-
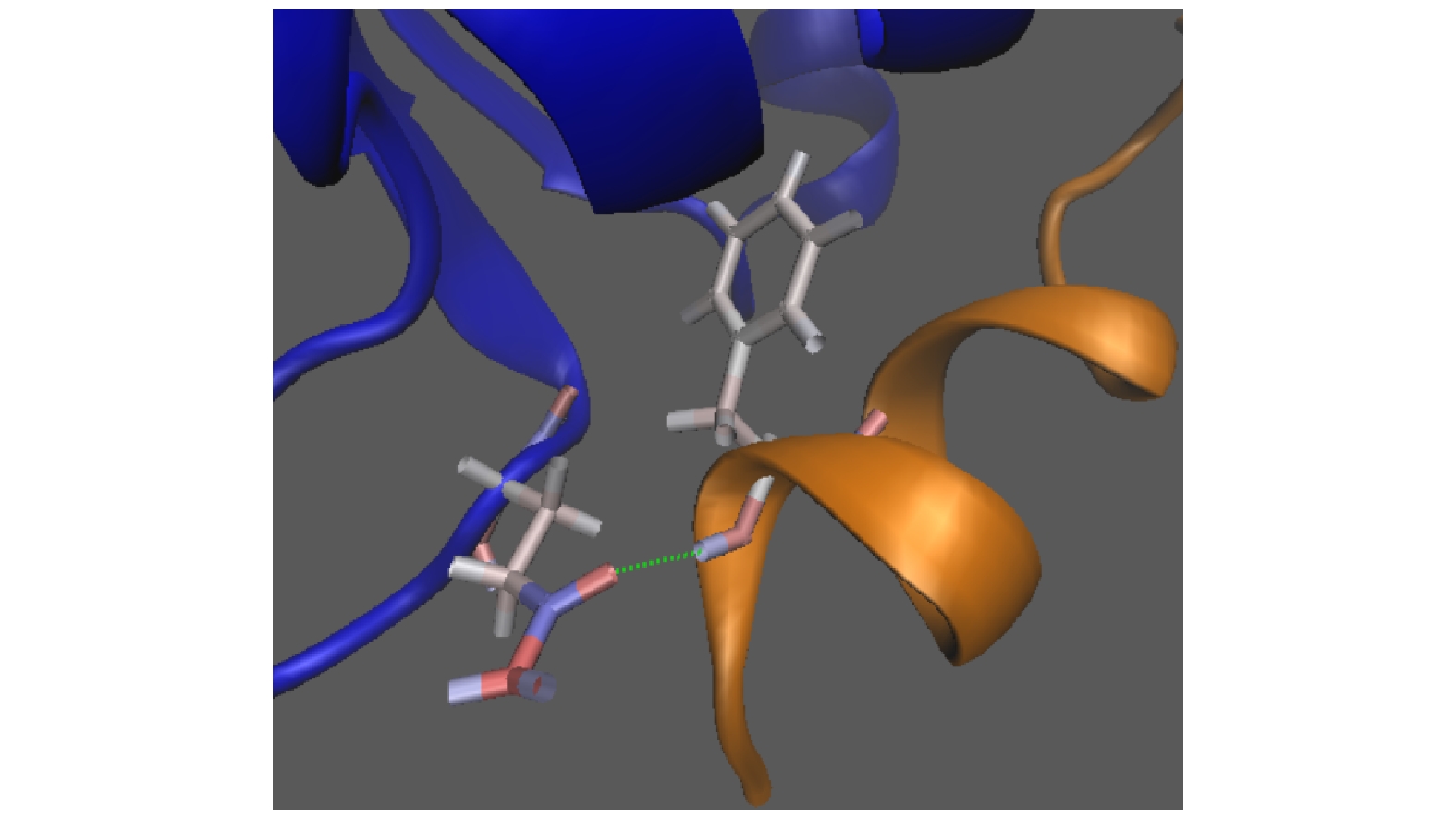
p53蛋白是一种与细胞周期停滞和细胞凋亡有关的蛋白质. 在受到细胞压力或环境扰动后, p53促进下游多个靶基因的转录, 介导肿瘤抑制. MDM2是主要的E3泛素连接酶, 也是p53的负调控因子. MDM2可促进p53的泛素化和核输出, 抑制p53的抑癌活性. 因此MDM2对p53的负调控始终是肿瘤治疗中急切需要解决的问题. Nutlin-3a是被证明可以有效抑制p53-MDM2相互作用的小分子抑制剂. 本文使用全原子分子动力学模拟, 研究Nutlin-3a对p53-MDM2复合物的稳定性的影响. 结果表明, 通过引起p53和MDM2间Phe19-Gln72的氢键和Glu17-Lys94的盐桥发生的断裂, Nutlin-3a可以削弱p53和MDM2间的相互作用. 我们的工作对Nutlin-3a小分子抑制剂的作用机制进行了说明, 揭示了抗癌药物Nutlin-3a介导的p53-MDM2复合物亲和力降低的分子机制, 并为针对p53蛋白的有效抗癌治疗提供了理论基础.
-
关键词:
- 治癌药物设计 /
- p53-MDM2复合物 /
- Nutlin-3a抑制剂分子 /
- 分子动力学模拟
P53 is well recognized to be a tumor suppressor protein. In response to the external stress or environmental perturbation, p53 can promote the transcription of various target genes downstream, thus regulating the cell cycle, apoptosis, DNA repair, and angiogenesis. However, the activation of p53 is further activated by another protein, MDM2, which negatively regulates the level of p53 inverse and thus reduces the activation of p53. This phenomenon is a novel potential and promising strategy for cancer therapy, i.e. restoring the activity of p53 pathway through the competitive inhibitors that can occupy the p53-binding site of MDM2 and thus inhibit the interaction between p53 and MDM2. Recently, various kinds of the inhibitors have been designed for this purpose. The Nutlin family is a group of well investigated inhibitors, which shows high efficiency for tumor suppression. Nutlin-3a mimics the MDM2-binding site of p53 essentially, and blocks the binding of MDM2 to p53. Once getting free from MDM2, p53 rapidly accumulates in the nuclei of cancer cells, the p53 target genes and the p53 pathway are activated, thereby resulting in cell-cycle arrest and apoptosis. In our previous papers, we investigated the competition mechanism between Nutlin3 and p53 in vitro by using molecular dynamics simulations. We found that Nutlin3 can bind faster than p53 to prevent p53 from binding to MDM2 when Nutlin-3a and p53 have equal distance from MDM2. Nutlin-3a can also bind to the p53-MDM2 complex to disturb and weaken the interactions between p53 and MDM2. However, the underlying mechanisms of p53-MDM2 complex instability in vivo are still unclear. And these inhibitors also have a variety of specificities and biological toxicities in vivo environment. In this study, we go a further step to investigate the effect of Nutlin-3a on the stability of p53-MDM2 complex in physiological environment with the aid of the molecular mechanics/generalized borne surface area (MM/GBSA) method. In our simulations, a group of Nutlin-3a molecules are randomly put around the p53 binding pocket of MDM2 in the initial stages to examine the dynamics among p53, MDM2 and the group of Nutlin-3a molecules and to analyze the underlying competition mechanism between Nutlin3 and p53 binding to pocket of MDM2. We find that Nutlin-3a can induce the centroid distance between p53 and MDM2 to increase. Importantly, we show that Nutlin-3a weakens the binding affinity of p53-MDM2 complex. Consistently, Nutlin-3a breaks a hydrogen bond between Phe19-Gln72 and a salt bridge between Glu17-Lys94, which weakens the interactions between p53 and MDM2. From the systematic biology point of view, the regulation of p53 by MDM2 is extremely sensitive to the strength of the p53-MDM2 interaction. The avianization of the interactions between p53 and MDM2 by Nutlin-3a can promote p53 to restore its suppression functions on tumor development. This study may be helpful in understanding the molecular mechanisms of p53-MDM2 complex instability mediated by Nutlin-3a and also in searching for the effective inhibitors of p53-MDM2 interaction. -
Keywords:
- cancer drug design /
- p53-MDM2 complex /
- Nutlin-3a inhibitor molecule /
- molecular dynamics simulation
[1] Levine A J 1997 Cell 88 323
 Google Scholar
Google Scholar
[2] Vogelstein B, Lane D, Levine A J 2000 Nature 408 307
 Google Scholar
Google Scholar
[3] Vousden K H, Lu X 2002 Nat. Rev. Cancer 2 594
 Google Scholar
Google Scholar
[4] Fridman J S, Lowe S W 2003 Oncogene 22 9030
 Google Scholar
Google Scholar
[5] Chen J 2016 CSH Perspect. Med. 6 a026104
 Google Scholar
Google Scholar
[6] Chène P 2003 Nat. Rev. Cancer 3 102
 Google Scholar
Google Scholar
[7] Vassilev L T, Vu B T, Graves B, et al. 2004 Science 303 844
 Google Scholar
Google Scholar
[8] Wiman K 2006 Cell Death Differ. 13 921
 Google Scholar
Google Scholar
[9] Hollstein M, Sidransky D, Vogelstein B, Harris C C 1991 Science 253 49
 Google Scholar
Google Scholar
[10] Batchelor E, Loewer A, Lahav G 2009 Nat. Rev. Cancer 9 371
 Google Scholar
Google Scholar
[11] Yan S, Zhuo Y 2006 Physica D 220 157
 Google Scholar
Google Scholar
[12] 张丽娟, 晏世伟, 卓益忠 2007 56 2442
 Google Scholar
Google Scholar
Zhang L J, Yan S W, Zhuo Y Z 2007 Acta Phys. Sin. 56 2442
 Google Scholar
Google Scholar
[13] Bond G L, Hu W, Bond E E, et al. 2004 Cell 119 591
 Google Scholar
Google Scholar
[14] Rayburn E, Zhang R, He J, Wang H 2005 Curr. Drug Targets 5 27
 Google Scholar
Google Scholar
[15] Michael D, Oren M 2003 Semin. Cancer Biol. 13 49
 Google Scholar
Google Scholar
[16] Gudkov A V 2004 Nat. Med. 10 1298
 Google Scholar
Google Scholar
[17] Grinkevich V, Issaeva N, Hossain S, Pramanik A, Selivanova G 2005 Nat. Med. 11 1136
 Google Scholar
Google Scholar
[18] Anifowose A, Agbowuro A A, Yang X, Wang B 2020 Med. Chem. Res. 29 1105
 Google Scholar
Google Scholar
[19] Zhu H, Gao H, Ji Y, Zhou Q, Du Z, Tian L, Jiang Y, Yao K, Zhou Z 2022 J. Hematol. Oncol. 15 1
 Google Scholar
Google Scholar
[20] Wang S, Zhao Y, Aguilar A, Bernard D, Yang C Y 2017 CSH Perspect. Med. 7 a026245
 Google Scholar
Google Scholar
[21] Nayak S K, Khatik G L, Narang R, Monga V, Chopra H K 2018 Curr. Drug Targets 18 749
 Google Scholar
Google Scholar
[22] Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P 1996 Science 274 948
 Google Scholar
Google Scholar
[23] Tovar C, Rosinski J, Filipovic Z, et al. 2006 PNAS 103 1888
 Google Scholar
Google Scholar
[24] Kojima K, Konopleva M, Samudio I J, et al. 2005 Blood 106 3150
 Google Scholar
Google Scholar
[25] Liu S X, Yan S W 2017 Chin. Phys. Lett. 34 118701
 Google Scholar
Google Scholar
[26] Liu S X, Geng Y Z, Yan S W 2017 Front. Phys. 12 1
 Google Scholar
Google Scholar
[27] Phillips J C, Hardy D J, Maia J D, Stone J E, Ribeiro J V, Bernardi R C, Buch R, Fiorin G, Hénin J, Jiang W, et al. 2020 J. Chem. Phys. 153 044130
 Google Scholar
Google Scholar
[28] Bai Q, Tan S, Xu T, Liu H, Huang J, Yao X 2021 Brief. Bioinform. 2 2
 Google Scholar
Google Scholar
[29] Suleman M, Yousafi Q, Ali J, et al. 2021 Comput. Biol. Med. 138 104936
 Google Scholar
Google Scholar
[30] Khan A, Zia T, Suleman M, Khan T, Ali S S, Abbasi A A, Mohammad A, Wei D Q 2021 J. Cell. Physiol. 236 7045
 Google Scholar
Google Scholar
[31] Khan A, Wei D Q, Kousar K, et al. 2021 Chembiochem 22 2641
 Google Scholar
Google Scholar
[32] Das P, Mattaparthi V S K 2020 ACS Omega 5 8449
 Google Scholar
Google Scholar
[33] Lev Bar-Or R, Maya R, Segel L A, Alon U, Levine A J, Oren M 2000 PNAS 97 11250
 Google Scholar
Google Scholar
-
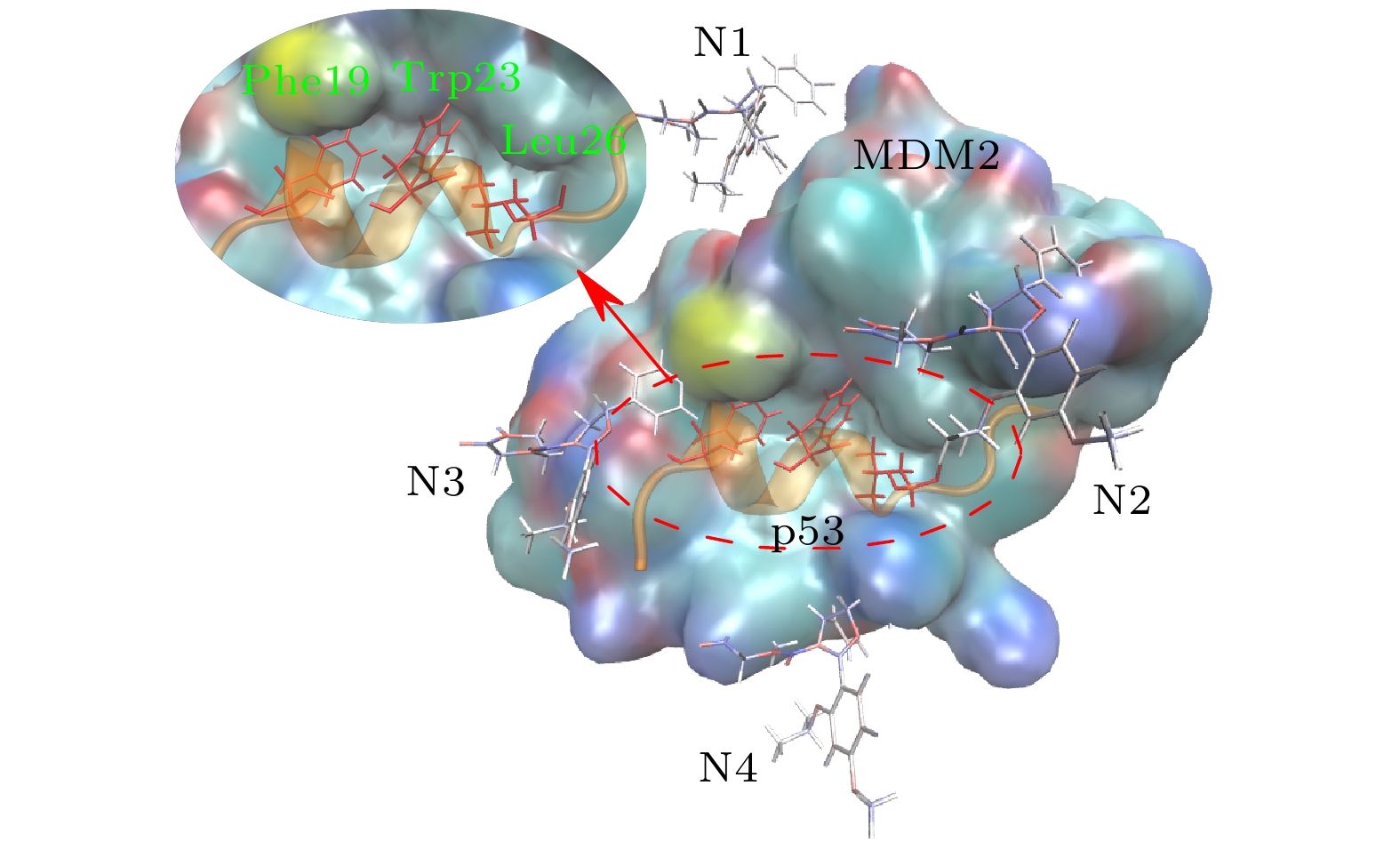
图 1 p53-MDM2复合物与4个Nutlin-3a的初始构象. MDM2用彩色表面表示; p53用橙色螺旋表示; 4个Nutlin-3a小分子由棒状结构给出(N1—N4), p53的3个残基Phe19, Trp23和Leu26插入到MDM2的疏水腔内, 用红色棒状结构表示
Fig. 1. Initial structure of p53-MDM2 complex with four Nutlin-3a. Colored surface shows MDM2; p53 helix is colored in orange. The small molecule shown by the stick is Nutlin-3a. Three amino acids (Phe19, Trp23 and Leu26) of p53 colored in red insert into the hydrophobic cavity of MDM2
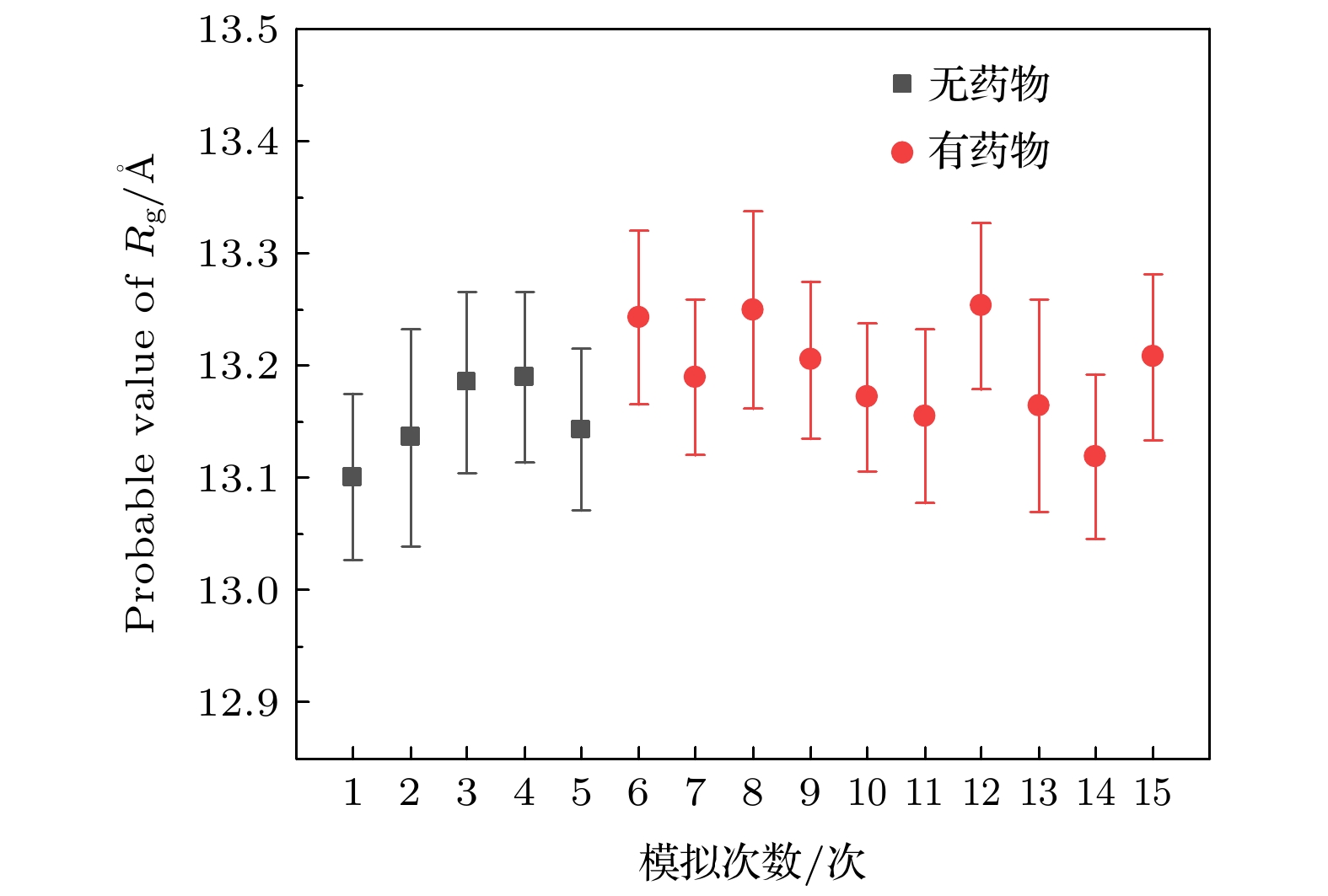
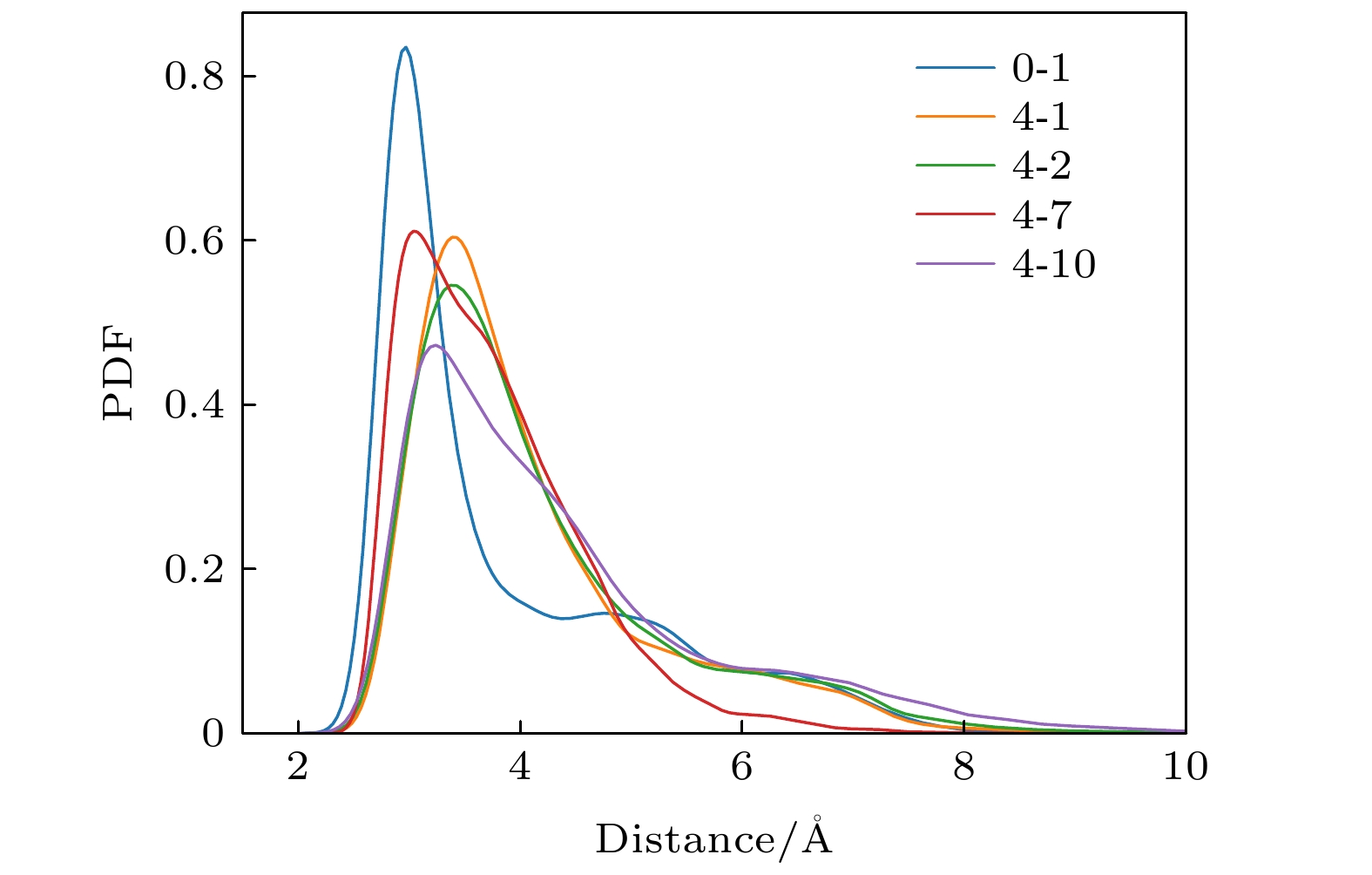
图 2 Nutlin-3a导致的p53-MDM2复合物构象变化. p53主链碳原子在(a)无药物体系和(b), (c) 有药物体系中相对于初始构象的RMSD演化. 0-1表示无药物体系的第1次模拟, 4-1表示有4个药物体系的第1次模拟, 以此类推. p53和MDM2的质心距离在(d)无药物体系和(e), (f)有药物体系中随时间的演化, 每次模拟的距离平均值在图中给出. (g)无药物体系(上)和有药物体系(中, 下)在4个不同模拟时间点的快照, p53用橙色螺旋表示, MDM2用蓝色表示. 这些数据表明, Nutlin-3a影响了p53和MDM2的结合构象
Fig. 2. Characterization of Nutlin-3a-induced structural changes of the MDM2-p53 complex. Time evolution of RMSD of p53 main-chain carbon atoms in (a) drug-free and (b), (c) drug-enabled system, the average RMSD of the five simulations is given in the figure. 0-1 represents the first simulation of a drug-free system, 4-1 represents the first simulation of a four-drug system, and so on. Time evolution of centroid distance between p53 and MDM2 in (d) drug-free system and (e), (f) drug-enabled system. The average distance of each simulation is given in the figure. (g) Representative snapshots of drug-free systems (top) and drug-enabled systems (middle, bottom) at four different simulation time points. p53 is represented by orange helix and MDM2 is represented by blue. These data reveal that Nutlin-3a affects the binding conformation of p53 and MDM2
表 1 p53-MDM2 的结合自由能(kcal/mol)
Table 1. Binding free energy of the p53-MDM2 complex
结合自由能/($\rm{kcal\cdot mol^{-1}}$) 无药物(5次模拟) 有药物(10次模拟) –47.0654 –44.92 –42.7475 –44.3723 –37.5379 –45.1035 –45.3874 –44.4957 –43.392 –45.0711 –48.7658 –45.6778 –47.8049 –41.1574 –34.5264 表 2 Phe19-Gln72间的氢键形成概率
Table 2. Probability of hydrogen bond formation between Phe19-Gln72
氢键形成概率/% 无药物(5次模拟) 有药物(10次模拟) 50.08 5.77 60.86 50.08 6.11 21.72 35.10 48.02 67.94 50.91 66.64 43.51 41.60 35.25 10.77 表 3 Lys94-Glu17 间盐桥的形成概率
Table 3. The probability of salt bridge formation between Lys94-Glu17
盐桥形成概率/% 无药物(5次模拟) 有药物(10次模拟) 37.77 0 42.21 33.62 2.81 5.03 26.90 21.22 20.43 50.32 77.16 10.8 19.07 35.25 0 -
[1] Levine A J 1997 Cell 88 323
 Google Scholar
Google Scholar
[2] Vogelstein B, Lane D, Levine A J 2000 Nature 408 307
 Google Scholar
Google Scholar
[3] Vousden K H, Lu X 2002 Nat. Rev. Cancer 2 594
 Google Scholar
Google Scholar
[4] Fridman J S, Lowe S W 2003 Oncogene 22 9030
 Google Scholar
Google Scholar
[5] Chen J 2016 CSH Perspect. Med. 6 a026104
 Google Scholar
Google Scholar
[6] Chène P 2003 Nat. Rev. Cancer 3 102
 Google Scholar
Google Scholar
[7] Vassilev L T, Vu B T, Graves B, et al. 2004 Science 303 844
 Google Scholar
Google Scholar
[8] Wiman K 2006 Cell Death Differ. 13 921
 Google Scholar
Google Scholar
[9] Hollstein M, Sidransky D, Vogelstein B, Harris C C 1991 Science 253 49
 Google Scholar
Google Scholar
[10] Batchelor E, Loewer A, Lahav G 2009 Nat. Rev. Cancer 9 371
 Google Scholar
Google Scholar
[11] Yan S, Zhuo Y 2006 Physica D 220 157
 Google Scholar
Google Scholar
[12] 张丽娟, 晏世伟, 卓益忠 2007 56 2442
 Google Scholar
Google Scholar
Zhang L J, Yan S W, Zhuo Y Z 2007 Acta Phys. Sin. 56 2442
 Google Scholar
Google Scholar
[13] Bond G L, Hu W, Bond E E, et al. 2004 Cell 119 591
 Google Scholar
Google Scholar
[14] Rayburn E, Zhang R, He J, Wang H 2005 Curr. Drug Targets 5 27
 Google Scholar
Google Scholar
[15] Michael D, Oren M 2003 Semin. Cancer Biol. 13 49
 Google Scholar
Google Scholar
[16] Gudkov A V 2004 Nat. Med. 10 1298
 Google Scholar
Google Scholar
[17] Grinkevich V, Issaeva N, Hossain S, Pramanik A, Selivanova G 2005 Nat. Med. 11 1136
 Google Scholar
Google Scholar
[18] Anifowose A, Agbowuro A A, Yang X, Wang B 2020 Med. Chem. Res. 29 1105
 Google Scholar
Google Scholar
[19] Zhu H, Gao H, Ji Y, Zhou Q, Du Z, Tian L, Jiang Y, Yao K, Zhou Z 2022 J. Hematol. Oncol. 15 1
 Google Scholar
Google Scholar
[20] Wang S, Zhao Y, Aguilar A, Bernard D, Yang C Y 2017 CSH Perspect. Med. 7 a026245
 Google Scholar
Google Scholar
[21] Nayak S K, Khatik G L, Narang R, Monga V, Chopra H K 2018 Curr. Drug Targets 18 749
 Google Scholar
Google Scholar
[22] Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P 1996 Science 274 948
 Google Scholar
Google Scholar
[23] Tovar C, Rosinski J, Filipovic Z, et al. 2006 PNAS 103 1888
 Google Scholar
Google Scholar
[24] Kojima K, Konopleva M, Samudio I J, et al. 2005 Blood 106 3150
 Google Scholar
Google Scholar
[25] Liu S X, Yan S W 2017 Chin. Phys. Lett. 34 118701
 Google Scholar
Google Scholar
[26] Liu S X, Geng Y Z, Yan S W 2017 Front. Phys. 12 1
 Google Scholar
Google Scholar
[27] Phillips J C, Hardy D J, Maia J D, Stone J E, Ribeiro J V, Bernardi R C, Buch R, Fiorin G, Hénin J, Jiang W, et al. 2020 J. Chem. Phys. 153 044130
 Google Scholar
Google Scholar
[28] Bai Q, Tan S, Xu T, Liu H, Huang J, Yao X 2021 Brief. Bioinform. 2 2
 Google Scholar
Google Scholar
[29] Suleman M, Yousafi Q, Ali J, et al. 2021 Comput. Biol. Med. 138 104936
 Google Scholar
Google Scholar
[30] Khan A, Zia T, Suleman M, Khan T, Ali S S, Abbasi A A, Mohammad A, Wei D Q 2021 J. Cell. Physiol. 236 7045
 Google Scholar
Google Scholar
[31] Khan A, Wei D Q, Kousar K, et al. 2021 Chembiochem 22 2641
 Google Scholar
Google Scholar
[32] Das P, Mattaparthi V S K 2020 ACS Omega 5 8449
 Google Scholar
Google Scholar
[33] Lev Bar-Or R, Maya R, Segel L A, Alon U, Levine A J, Oren M 2000 PNAS 97 11250
 Google Scholar
Google Scholar
计量
- 文章访问数: 5840
- PDF下载量: 80
- 被引次数: 0













 下载:
下载: