-
过渡态是物理化学家理解和调控生物大分子相关功能微观机制的关键. 因其存在时间极短, 难以被实验手段捕捉, 全面刻画其结构必须通过物理定律驱动的模拟计算搜索予以实现. 然而, 与化学反应过程只涉及少量原子不同, 生物大分子的功能性构象变化所涉的原子和坐标数量巨大, 搜索其过渡态将不可避免地遭遇维数灾难, 即反应坐标问题, 因而催生了多种应对策略和算法. 同时, 随着近年来新型机器学习算法的大量涌现和日臻成熟, 融入机器学习范式的过渡态搜索算法也已出现. 本文首先回顾和梳理过渡态搜索代表性算法的设计思想, 包括依赖集合变量的温和爬升动力学(gentlest ascent dynamics, GAD)、有限温度弦方法(finite temperature string, FTS)、快速断层扫描法(fast tomographic)、基于旅行商的自动路径搜索算法TAPS, 以及过渡路径采样法(transition path sampling, TPS). 然后, 重点介绍TPS与强化学习融合而成的新型路径采样算法, 解析强化学习在其中的作用, 并厘清其适用场景. 最后, 我们提出一种将降维算法与GAD深度融合的新构想, 讨论研发可保留过渡态信息的新型降维算法的必要性及可行性.Transition state is a key concept for chemists to understand and fine-tune the conformational changes of large biomolecules. Due to its short residence time, it is difficult to capture a transition state via experimental techniques. Characterizing transition states for a conformational change therefore is only achievable via physics-driven molecular dynamics simulations. However, unlike chemical reactions which involve only a small number of atoms, conformational changes of biomolecules depend on numerous atoms and therefore the number of their coordinates in our 3D space. The searching for their transition states will inevitably encounter the curse of dimensionality, i.e. the reaction coordinate problem, which invokes the invention of various algorithms for solution. Recent years, new machine learning techniques and the incorporation of some of them into the transition state searching methods emerged. Here, we first review the design principle of representative transition state searching algorithms, including the collective-variable (CV)-dependent gentlest ascent dynamics, finite temperature string, fast tomographic, travelling-salesman based automated path searching, and the CV-independent transition path sampling. Then, we focus on the new version of TPS that incorporates reinforcement learning for efficient sampling, and we also clarify the suitable situation for its application. Finally, we propose a new paradigm for transition state searching, a new dimensionality reduction technique that preserves transition state information and combines gentlest ascent dynamics.
-
Keywords:
- transition state /
- gentlest ascent dynamics /
- path methods /
- reinforcement learning /
- generative models
[1] Edman L, Földes-Papp Z, Wennmalm S, Rigler R 1999 Chem. Phys. 247 11
 Google Scholar
Google Scholar
[2] Evenäs J, Malmendal A, Thulin E, Carlstrӧm G, Forsén S 1998 Biochemistry 37 13744
 Google Scholar
Google Scholar
[3] Hanson J A, Duderstadt K, Watkins L P, Bhattacharyya S, Brokaw J B, Chu J W, Yang H 2007 Proc. Natl. Acad. Sci. USA 104 18055
 Google Scholar
Google Scholar
[4] Moffat K 1989 Annu. Rev. Biophys. Chem. 18 309
 Google Scholar
Google Scholar
[5] Huang C, Kalodimos C G 2017 Annu. Rev. Biophys. 46 317
 Google Scholar
Google Scholar
[6] Weissenberger G, Henderikx R J M, Peters P J 2021 Nat. Methods 18 463
 Google Scholar
Google Scholar
[7] Clegg R M 1995 Curr. Opin. Biotechnol. 6 103
 Google Scholar
Google Scholar
[8] Karplus M, McCammon J A 2002 Nat. Struct. Biol. 9 646
 Google Scholar
Google Scholar
[9] Hollingsworth S A, Dror R O 2018 Neuron 99 1129
 Google Scholar
Google Scholar
[10] Bernèche S, Roux B 2001 Nature 414 73
 Google Scholar
Google Scholar
[11] Khafizov K, Perez C, Koshy C, Quick M, Fendler K, Ziegler C, Forrest L R 2012 Proc. Natl. Acad. Sci. USA 109 E3035
 Google Scholar
Google Scholar
[12] Li J, Shaikh S A, Enkavi G, Wen P C, Huang Z, Tajkhorshid E 2013 Proc. Natl. Acad. Sci. USA 110 7696
 Google Scholar
Google Scholar
[13] Dror, R O, Green H F, Valant C, Borhani D W, Valcourt J R, Pan A C, Arlow D H, Canals M, Lane J R, Rahmani R, Baell J B, Sexton P M, Christopoulos A, Shaw D E 2013 Nature 503 295
 Google Scholar
Google Scholar
[14] Wacker D, Stevens R C, Roth B L 2017 a Cell 170 414
 Google Scholar
Google Scholar
[15] Wacker D, Wang S, McCorvy J D, Betz R M, Venkatakrishnan A J, Levit A, Lansu K, Schools Z L, Che T, Nichols D E, Dror R O, Roth B L 2017 Cell 168 377
 Google Scholar
Google Scholar
[16] McCorvy J D, Butler K V, Kelly B, Rechsteiner K, Karpiak J, Betz R M, Kormos B L, Shoichet B K, Dror R O, Jin J, Roth B L 2018 Nat. Chem. Biol. 14 126
 Google Scholar
Google Scholar
[17] Provasi D, Artacho M C, Negri A, Mobarec J C, Filizola M 2011 PLoS Comput. Biol. 7 e1002193
 Google Scholar
Google Scholar
[18] Cordero-Morales J F, Jogini V, Lewis A, Vásquez V, Cortes D M, Roux B, Perozo E 2007 Nat. Struct. Mol. Biol. 14 1062
 Google Scholar
Google Scholar
[19] Fields J B, Németh-Cahalan K L, Freites J A, Vorontsova I, Hall J E, Tobias D J 2017 J. Biol. Chem. 292 185
 Google Scholar
Google Scholar
[20] Groban E S, Narayanan A, Jacobson M P 2006 PLoS Comput. Biol. 2 e32
 Google Scholar
Google Scholar
[21] Liu Y, Ke M, Gong H 2015 Biophys. J. 109 542
 Google Scholar
Google Scholar
[22] Delemotte L, Tarek M, Klein M L, Amaral C, Treptow W 2011 Proc. Natl. Acad. Sci. USA 108 6109
 Google Scholar
Google Scholar
[23] Lindorff-Larsen K, Piana S, Dror R O, Shaw D E 2011 Science 334 517
 Google Scholar
Google Scholar
[24] Snow C D, Nguyen H, Pande V S, Gruebele M 2002 Nature 420 102
 Google Scholar
Google Scholar
[25] Dror R O, Arlow D H, Maragakis P, Mildorf T J, Pan A C, Xu H, Borhani D W, Shaw D E 2011 Proc. Natl. Acad. Sci. USA 108 18684
 Google Scholar
Google Scholar
[26] Dror R O, Pan A C, Arlow D H, Borhani D W, Maragakis P, Shan Y, Xu H, Shaw D E 2011 Proc. Natl. Acad. Sci. USA 108 13118
 Google Scholar
Google Scholar
[27] Gu Y, Shrivastava I H, Amara S G, Bahar I 2009 Proc. Natl. Acad. Sci. USA 106 2589
 Google Scholar
Google Scholar
[28] Latorraca N R, Fastman N M, Venkatakrishnan A J, Frommer W B, Dror R O, Feng L 2017 Cell 169 96
 Google Scholar
Google Scholar
[29] Stelzl L S, Fowler P W, Sansom M S, Beckstein O 2014 J. Mol. Biol. 426 735
 Google Scholar
Google Scholar
[30] Buch I, Giorgino T, De Fabritiis G 2011 Proc. Natl. Acad. Sci. USA 108 10184
 Google Scholar
Google Scholar
[31] Liang R, Swanson J M J, Madsen J J, Hong M, DeGrado W F, Voth G A 2016 Proc. Natl. Acad. Sci. USA 113 E6955
 Google Scholar
Google Scholar
[32] Suomivuori C M, Gamiz-Hernandez A P, Sundholm D, Kaila V R I 2017 Proc. Natl. Acad. Sci. USA 114 7043
 Google Scholar
Google Scholar
[33] Tajkhorshid E, Nollert P, Jensen M Ø, Miercke L J W, O’Connell J, Stroud R M, Schulten K 2002 Science 296 525
 Google Scholar
Google Scholar
[34] Watanabe A, Choe S, Chaptal V, Rosenberg J M, Wright E M, Grabe M, Abramson J 2010 Nature 468 988
 Google Scholar
Google Scholar
[35] Dedmon M M, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson C M 2005 J. Am. Chem. Soc. 127 476
 Google Scholar
Google Scholar
[36] Nguyen H D, Hall C K 2004 Proc. Natl. Acad. Sci. USA 101 16180
 Google Scholar
Google Scholar
[37] Levitt M 1983 J. Mol. Biol. 168 595
 Google Scholar
Google Scholar
[38] Sugita Y, Okamoto Y 1999 Chem. Phys. Lett. 314 141
 Google Scholar
Google Scholar
[39] Rhee Y M, Pande V S 2003 Biophys. J. 84 775
 Google Scholar
Google Scholar
[40] Zhang W, Wu C, Duan Y 2005 J. Chem. Phys. 123 154105
 Google Scholar
Google Scholar
[41] Zhou R 2006 Protein Folding Protocols (Humana Totowa, NJ: Springer) pp205−223
[42] Sindhikara D, Meng Y, Roitberg A E 2008 J. Chem. Phys. 128 024103
 Google Scholar
Google Scholar
[43] Buchete N V, Hummer G 2008 Phys. Rev. E 77 030902
 Google Scholar
Google Scholar
[44] Rosta E, Hummer G 2009 J. Chem. Phys. 131 165102
 Google Scholar
Google Scholar
[45] Stelzl L S, Hummer G 2017 J. Chem. Theory Comput. 13 3927
 Google Scholar
Google Scholar
[46] Yang L J, Gao Q Y 2009 J. Chem. Phys. 131 214109
 Google Scholar
Google Scholar
[47] Yang L, Liu C W, Shao Q, Zhang J, Gao Y Q 2015 Acc. Chem. Res. 48 947
 Google Scholar
Google Scholar
[48] Yang Y I, Zhang J, Che X, Yang L J, Gao Y Q 2016 J. Chem. Phys. 144 094105
 Google Scholar
Google Scholar
[49] Yang Y I, Shao Q, Zhang J, Yang L J, Gao Y Q 2019 J. Chem. Phys. 151 070902
 Google Scholar
Google Scholar
[50] Huber T, Torda A E, Van Gunsteren W F 1994 J. Comput. -Aided Mol. Des. 8 695
 Google Scholar
Google Scholar
[51] Wada T, Kuroda K, Yoshida Y, Ogasawara K, Ogawa A, Endo S 2006 Neurosurg. Rev. 29 242
 Google Scholar
Google Scholar
[52] Hansen H S, Hünenberger P H 2010 J. Comput. Chem. 31 1
 Google Scholar
Google Scholar
[53] Perić-Hassler L, Hansen H S, Baron R, Hünenberger P H 2010 Carbohydr. Res. 345 1781
 Google Scholar
Google Scholar
[54] Grubmüller H 1995 Phys. Rev. E 52 2893
 Google Scholar
Google Scholar
[55] Schulze B G, Grubmüller H, Evanseck J D 2000 J. Am. Chem. Soc. 122 8700
 Google Scholar
Google Scholar
[56] Bouvier B, Grubmüller H 2007 Biophys. J. 93 770
 Google Scholar
Google Scholar
[57] Barducci A, Bonomi M, Parrinello M 2011 WIREs Comput. Mol. Sci. 1 826
 Google Scholar
Google Scholar
[58] Tiwary P, Parrinello M 2013 Phys. Rev. Lett. 111 230602
 Google Scholar
Google Scholar
[59] Bussi G, Laio A 2020 Nat. Rev. Phys. 2 200
 Google Scholar
Google Scholar
[60] Miao Y, Feher V A, McCammon J A 2015 J. Chem. Theory Comput. 11 3584
 Google Scholar
Google Scholar
[61] Miao Y, McCammon J A 2017 Annu. Rep. Comput. Chem. 13 231
 Google Scholar
Google Scholar
[62] Wang J, Arantes P R, Bhattarai A, et al. 2021 Wiley Interdiscip. Rev.: Comput. Mol. Sci. 11 e1521
 Google Scholar
Google Scholar
[63] Naritomi Y, Sotaro F 2011 J. Chem. Phys. 134 065101
 Google Scholar
Google Scholar
[64] Schwantes C R, Vijay S P 2013 J. Chem. Theory Comput. 9 2000
 Google Scholar
Google Scholar
[65] Perez-Hernandez G, Paul F, Giorgino T, Fabritiis G D, Noé F 2013 J. Chem. Phys. 139 015102
 Google Scholar
Google Scholar
[66] Bowman G R, Huang X H, Pande V S 2009 Methods 49 197
 Google Scholar
Google Scholar
[67] Metzner P, Noé F, Schütte C 2009 Phys. Rev. E 80 021106
 Google Scholar
Google Scholar
[68] Pande V S, Beauchamp K A, Bowman G R 2010 Methods 52 99
 Google Scholar
Google Scholar
[69] Prinz J H, Wu H, Sarich M, Keller B, Senne M, Held M, Chodera J D, Schütte C, Noé F 2011 J. Chem. Phys. 134 174105
 Google Scholar
Google Scholar
[70] Kellogg E H, Lange O F, Baker D 2012 J. Phys. Chem. B 116 11405
 Google Scholar
Google Scholar
[71] Yao Y, Cui R Z, Bowman G R, Silva D A, Sun J, Huang X H 2013 J. Chem. Phys. 138 174106
 Google Scholar
Google Scholar
[72] McGibbon R T, Schwantes C R, Pande V S 2014 J. Phys. Chem. B 118 6475
 Google Scholar
Google Scholar
[73] Nuske F, Keller B G, Pérez-Hernández G, Mey A S J, Noé F 2014 J. Chem. Theory Comput. 10 1739
 Google Scholar
Google Scholar
[74] Sheong F K, Silva D A, Meng L, Zhao Y, Huang X H 2015 J. Chem. Theory Comput. 11 17
 Google Scholar
Google Scholar
[75] Zhu L Z, Sheong F K, Zeng X, Huang X H 2016 Phys. Chem. Chem. Phys. 18 30228
 Google Scholar
Google Scholar
[76] Wang W, Cao S, Zhu L Z, Huang X H 2018 WIREs Comput. Mol. Sci. 8 e1343
 Google Scholar
Google Scholar
[77] Husic B E, Pande V S 2018 J. Am. Chem. Soc. 140 2386
 Google Scholar
Google Scholar
[78] Konovalov K A, Unarta I C, Cao S, Goonetilleke E C, Huang X H 2021 J. Am. Chem. Soc. Au. 1 1330
 Google Scholar
Google Scholar
[79] E W, Zhou X 2011 Nonlinearity 24 1831
 Google Scholar
Google Scholar
[80] Samanta A, Chen M, Yu T Q, Tuckerman M E 2014 J. Chem. Phys. 140 164109
 Google Scholar
Google Scholar
[81] Chen M, Yu T Q, Tuckerman M E 2015 Proc. Natl. Acad. Sci. USA 112 3235
 Google Scholar
Google Scholar
[82] E W, Ren W, Vanden-Eijnden E 2002 Phys. Rev. B 66 052301
 Google Scholar
Google Scholar
[83] E W, Ren W, Vanden-Eijnden E 2005 J. Phys. Chem. B 109 6688
 Google Scholar
Google Scholar
[84] Maragliano L, Fischer A, Vanden-Eijnden E, Ciccotti G 2006 J. Chem. Phys. 125 024106
 Google Scholar
Google Scholar
[85] Ren W, Vanden-Eijnden E 2007 J. Chem. Phys. 126 164103
 Google Scholar
Google Scholar
[86] Maragliano L, Vanden-Eijnden E 2007 Chem. Phys. Lett. 446 182
 Google Scholar
Google Scholar
[87] Pan A C, Sezer D, Roux B 2008 J. Phys. Chem. B 112 3432
 Google Scholar
Google Scholar
[88] Chen C, Huang Y, Xiao Y 2012 Phys. Rev. E 86 031901
 Google Scholar
Google Scholar
[89] Chen C, Huang Y, Ji X, Xiao Y 2013 J. Chem. Phys. 138 164122
 Google Scholar
Google Scholar
[90] Chen C J, Huang Y Z, Jiang X W, Xiao Y 2014 J. Chem. Phys. 141 154109
 Google Scholar
Google Scholar
[91] Zhu L Z, Sheong F K, Cao S, Liu S, Unarta I C, Huang X H 2019 J. Chem. Phys. 150 124105
 Google Scholar
Google Scholar
[92] Xi K, Hu Z, Wu Q, Wei M, Qian R, Zhu L Z 2021 J. Chem. Theory Comput. 17 5301
 Google Scholar
Google Scholar
[93] Wang L, Xi K, Zhu L Z, Da L T 2022 J. Chem. Inf. Model. 62 3213
 Google Scholar
Google Scholar
[94] Xi K, Zhu L Z 2022 Int. J. Mol. Sci. 23 14628
 Google Scholar
Google Scholar
[95] Xi K, Zhu L Z 2023 A Practical Guide to Recent Advances in Multiscale Modeling and Simulation of Biomolecules (AIP Publishing) pp9-1−9-24
[96] Vanden-Eijnden E 2006 Computer Simulations in Condensed Matter: From Materials to Chemical Biology (Berlin: Springer) pp453−493
[97] Vanden-Eijnden E 2010 Annu. Rev. Phys. Chem. 61 391
 Google Scholar
Google Scholar
[98] Dellago C, Bolhuis P G, Csajka F S, Chandler D 1998 J. Chem. Phys. 108 1964
 Google Scholar
Google Scholar
[99] Bolhuis P G, Dellago C, Chandler D 1998 Faraday Discuss. 110 421
 Google Scholar
Google Scholar
[100] Dellago C, Bolhuis P G, Chandler D 1999 J. Chem. Phys. 110 6617
 Google Scholar
Google Scholar
[101] Dellago C, Bolhuis P G, Geissler P L 2002 Adv. Chem. Phys. 123 1
 Google Scholar
Google Scholar
[102] Noé F, Tkatchenko A, Müller K R, Clementi C 2020 Annu. Rev. Phys. Chem. 71 361
 Google Scholar
Google Scholar
[103] AlQuraishi M, Sorger P K 2021 Nat. Methods 18 1169
 Google Scholar
Google Scholar
[104] Karniadakis G E, Kevrekidis I G, Lu L, Perdikaris P, Wang S, Yang L 2021 Nat. Rev. Phys. 3 422
 Google Scholar
Google Scholar
[105] Ju F, Zhu J, Shao B, Kong L, Liu T Y, Zheng W M, Bu D 2021 Nat. Commum. 12 2535
 Google Scholar
Google Scholar
[106] Huang B, Xu Y, Hu X H, Liu Y R, Liao S H, Zhang J H, Huang C D, Hong J J, Chen Q, Liu H Y 2022 Nature 602 523
 Google Scholar
Google Scholar
[107] Du Z, Su H, Wang W, Ye L, Wei H, Peng Z, Anishchenko I, Baker D, Yang J 2021 Nat. Prot. 16 5634
 Google Scholar
Google Scholar
[108] Dai M, Dong Z, Xu K, Zhang Q C 2023 J. Mol. Biol. 435 168059
 Google Scholar
Google Scholar
[109] Yuan Q M, Chen J W, Zhao H Y, Zhou Y Q, Yang Y D 2022 Bioinformatics 38 125
 Google Scholar
Google Scholar
[110] Su M Y, Feng G, Liu Z, Li Y, Wang R 2020 J. Chem. Inf. Model. 60 1122
 Google Scholar
Google Scholar
[111] Zeng C, Jian Y, Vosoughi S, Zeng C, Zhao Y 2023 Nat. Commum. 14 1060
 Google Scholar
Google Scholar
[112] Noé F, Olsson S, Köhler J, Wu H 2019 Science 365 6457
 Google Scholar
Google Scholar
[113] Jung H, Covino R, Arjun A, Leitold C, Dellago C, Bolhuis P G, Hummer G 2023 Nat. Comput. Sci. 3 334
 Google Scholar
Google Scholar
[114] Weiss D R, Levitt M 2009 J. Mol. Biol. 385 665
 Google Scholar
Google Scholar
[115] Isralewitz B, Gao M, Schulten K 2001 Curr. Opin. Struct. Biol. 11 224
 Google Scholar
Google Scholar
[116] Schlitter J, Engels M, Krüger P 1994 J. Mol. Graph. 12 84
 Google Scholar
Google Scholar
[117] Torrie G M, Valleau J P 1977 J. Comput. Phys. 23 187
 Google Scholar
Google Scholar
[118] Ryckaert J P, Ciccotti G, Berendsen H J 1977 J. Comput. Phys. 23 327
 Google Scholar
Google Scholar
[119] Babin V, Roland C, Sagui C 2008 J. Chem. Phys. 128 134101
 Google Scholar
Google Scholar
[120] Branduardi D, Gervasio F L, Parrinello M 2007 J. Chem. Phys. 126 054103
 Google Scholar
Google Scholar
[121] Applegate D L, Bixby R E, Chvátal V, Cook W J 2011 The Traveling Salesman Problem: A Computational Study (Princeton University Press) pp1−58
[122] Cox M A A, Cox T F 2008 Handbook of Data Visualization (Berlin: Springer) pp315−347
[123] Barducci A, Bussi G, Parrinello M 2008 Phys. Rev. Lett. 100 020603
 Google Scholar
Google Scholar
[124] Fischmann T O, Smith C K, Mayhood T W, Myers J E, Reichert J P, Mannarino A, Carr D, Zhu H, Wong J, Yang R S, Le H V, Madison V S 2009 Biochemistry 48 2661
 Google Scholar
Google Scholar
[125] Hanrahan A J, Sylvester B E, Chang M T, et al. 2020 Cancer Res. 80 4233
 Google Scholar
Google Scholar
[126] Schmidt M, Lipson H 2009 Science 324 81
 Google Scholar
Google Scholar
[127] Jung H, Okazaki K, Hummer G 2017 J. Chem. Phys. 147 152716
 Google Scholar
Google Scholar
[128] Swenson D W H, Prinz J H, Noe F, Chodera J D, Bolhuis P G 2019 J. Chem. Theory Comput. 15 813
 Google Scholar
Google Scholar
[129] Swenson D W H, Prinz J H, Noe F, Chodera J D, Bolhuis P G 2019 J. Chem. Theory Comput. 15 837
 Google Scholar
Google Scholar
[130] Glielmo A, Husic B E, Rodriguez A, Clementi C, Noé F, Laio A 2021 Chem. Rev. 121 9722
 Google Scholar
Google Scholar
-
图 1 (a)依赖集合变量的过渡态搜索示意图, 需由生物分子(以丙戊酸二肽为例)体系所在的高维相空间(phase space)选取少量集合变量CV强行“定向降维”, 后在此低维CV空间利用非路径类方法或路径方法, 找到过渡态(Transition State), 并给出微观机制解释(mechanism interpretation); (b)非路径类的GAD算法原理示意图; (c), (d)两类路径类搜索算法原理示意图
Fig. 1. (a) Illustration of the flow-chart of the collective variables (CVs) based transition state searching. A low dimensional space must be constructed with the CVs, which are arbitrary a priori guess about the mechanism. The transition state(s) is then determined by either the non-path or path methods. (b) The non-path method GAD. Path methods of (c) finite temperature string and (d) fast tomographic.
图 2 (a) PCV构建[120]和TAPS Method[91–95,121]算法原理示意图; (b)基于伞形采样方法得到的TAPS算法确定的MEK1由Loop-Out到达Loop-In转变过程最小自由能路径(MFEP)的自由能图景及相应的微观转变机制[92]
Fig. 2. (a) Illustration for the construction of PCV and the flow-chart of the TAPS method; (b) TAPS revealed the free energy landscape and the transition states for the transition from the Loop-Out state of MEK1 to its Loop-In state[92].
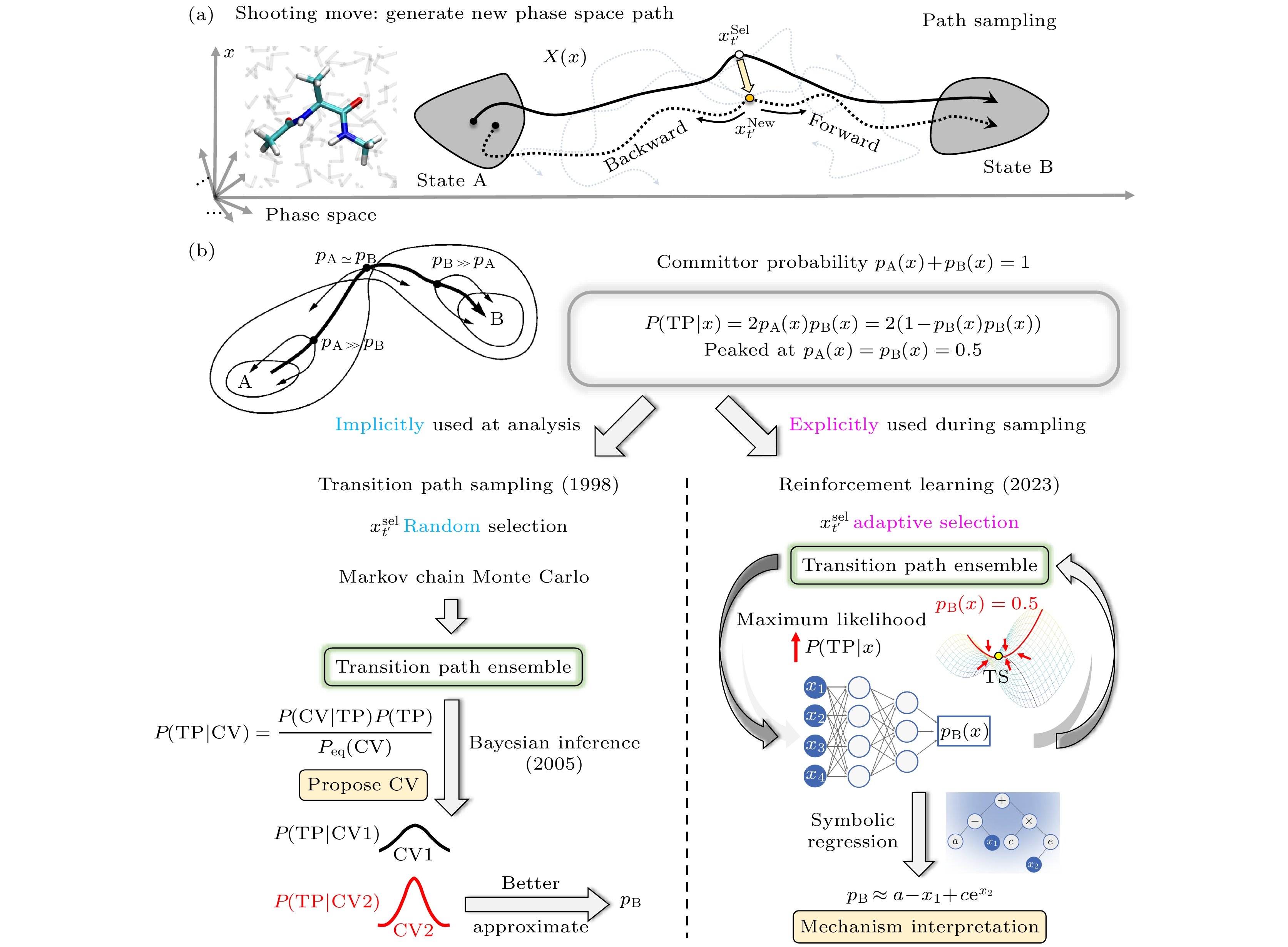
图 3 路径采样算法的基本原理示意图 (a)路径采样中生成新相空间路径的shooting move; (b)传统过渡路径采样(左侧)的随机蒙特卡罗采样与过渡态分析原理[98–101], 融合强化学习的路径采样(右侧)在学习过程中不断促进采样起始点选择向过渡态集中[113]
Fig. 3. Schematics of path sampling methods. (a) Shooting move: select a phase space point on the current path, make a small perturbation to this point (redraw random initial velocities) and perform a set of simulations. (b) Path sampling is built upon the committor probability $ {p}_{{\mathrm{B}}} $. The traditional transition path sampling (left)[98–101] selects shooting points randomly and uses Monte Carlo for sampling; the transition state is characterized through post-analysis: choosing the CVs with the highest and narrowest distribution of P(TP|CV); the new reinforcement path sampling (right)[113] chooses shooting points adaptively and directly learns the committor probability $ {p}_{{\mathrm{B}}} $ with maximized P(TP|x). Symbolic regression of $ {p}_{{\mathrm{B}}} $ is used for mechanism interpretation.
图 4 物理化学家需要怎样的降维算法 (a)现有降维算法范式不保留过渡态信息, 不利于机制解析; (b)可能的替代范式, 基于生成模型研发可保留过渡态信息的可逆降维算法, 并与低维空间搜索过渡态的GAD联用
Fig. 4. Requirements on dimensionality reduction algorithms by physical chemists. (a) Current paradigm for dimensionality reduction and the main difficulties for the transition state searching. (b) Proposed alternative paradigm for transition state searching: combine dimensionality reduction that preserves transition state information with GAD.
表 1 主要过渡态搜索算法的总结分类
Table 1. Classification of the algorithms for transition state searching.
过渡态搜索算法分类 代表性算法 参考文献 备注 传统方法 依赖CV Gentlest ascent dynamics (GAD) [79—81] 非路径方法 Finite temperature string [82—87]
路径方法预设低维 Fast tomographic [88—90] 空间搜索 基于旅行商的路径搜索 TAPS [91—95] 融合AI 不依赖CV
高维空间搜索Transition path sampling [98—101] Reinforcement path sampling [113] 保留过渡态
信息的降维
低维空间搜索融合生成模型及GAD
的过渡态搜索(待研发)无 非路径方法 -
[1] Edman L, Földes-Papp Z, Wennmalm S, Rigler R 1999 Chem. Phys. 247 11
 Google Scholar
Google Scholar
[2] Evenäs J, Malmendal A, Thulin E, Carlstrӧm G, Forsén S 1998 Biochemistry 37 13744
 Google Scholar
Google Scholar
[3] Hanson J A, Duderstadt K, Watkins L P, Bhattacharyya S, Brokaw J B, Chu J W, Yang H 2007 Proc. Natl. Acad. Sci. USA 104 18055
 Google Scholar
Google Scholar
[4] Moffat K 1989 Annu. Rev. Biophys. Chem. 18 309
 Google Scholar
Google Scholar
[5] Huang C, Kalodimos C G 2017 Annu. Rev. Biophys. 46 317
 Google Scholar
Google Scholar
[6] Weissenberger G, Henderikx R J M, Peters P J 2021 Nat. Methods 18 463
 Google Scholar
Google Scholar
[7] Clegg R M 1995 Curr. Opin. Biotechnol. 6 103
 Google Scholar
Google Scholar
[8] Karplus M, McCammon J A 2002 Nat. Struct. Biol. 9 646
 Google Scholar
Google Scholar
[9] Hollingsworth S A, Dror R O 2018 Neuron 99 1129
 Google Scholar
Google Scholar
[10] Bernèche S, Roux B 2001 Nature 414 73
 Google Scholar
Google Scholar
[11] Khafizov K, Perez C, Koshy C, Quick M, Fendler K, Ziegler C, Forrest L R 2012 Proc. Natl. Acad. Sci. USA 109 E3035
 Google Scholar
Google Scholar
[12] Li J, Shaikh S A, Enkavi G, Wen P C, Huang Z, Tajkhorshid E 2013 Proc. Natl. Acad. Sci. USA 110 7696
 Google Scholar
Google Scholar
[13] Dror, R O, Green H F, Valant C, Borhani D W, Valcourt J R, Pan A C, Arlow D H, Canals M, Lane J R, Rahmani R, Baell J B, Sexton P M, Christopoulos A, Shaw D E 2013 Nature 503 295
 Google Scholar
Google Scholar
[14] Wacker D, Stevens R C, Roth B L 2017 a Cell 170 414
 Google Scholar
Google Scholar
[15] Wacker D, Wang S, McCorvy J D, Betz R M, Venkatakrishnan A J, Levit A, Lansu K, Schools Z L, Che T, Nichols D E, Dror R O, Roth B L 2017 Cell 168 377
 Google Scholar
Google Scholar
[16] McCorvy J D, Butler K V, Kelly B, Rechsteiner K, Karpiak J, Betz R M, Kormos B L, Shoichet B K, Dror R O, Jin J, Roth B L 2018 Nat. Chem. Biol. 14 126
 Google Scholar
Google Scholar
[17] Provasi D, Artacho M C, Negri A, Mobarec J C, Filizola M 2011 PLoS Comput. Biol. 7 e1002193
 Google Scholar
Google Scholar
[18] Cordero-Morales J F, Jogini V, Lewis A, Vásquez V, Cortes D M, Roux B, Perozo E 2007 Nat. Struct. Mol. Biol. 14 1062
 Google Scholar
Google Scholar
[19] Fields J B, Németh-Cahalan K L, Freites J A, Vorontsova I, Hall J E, Tobias D J 2017 J. Biol. Chem. 292 185
 Google Scholar
Google Scholar
[20] Groban E S, Narayanan A, Jacobson M P 2006 PLoS Comput. Biol. 2 e32
 Google Scholar
Google Scholar
[21] Liu Y, Ke M, Gong H 2015 Biophys. J. 109 542
 Google Scholar
Google Scholar
[22] Delemotte L, Tarek M, Klein M L, Amaral C, Treptow W 2011 Proc. Natl. Acad. Sci. USA 108 6109
 Google Scholar
Google Scholar
[23] Lindorff-Larsen K, Piana S, Dror R O, Shaw D E 2011 Science 334 517
 Google Scholar
Google Scholar
[24] Snow C D, Nguyen H, Pande V S, Gruebele M 2002 Nature 420 102
 Google Scholar
Google Scholar
[25] Dror R O, Arlow D H, Maragakis P, Mildorf T J, Pan A C, Xu H, Borhani D W, Shaw D E 2011 Proc. Natl. Acad. Sci. USA 108 18684
 Google Scholar
Google Scholar
[26] Dror R O, Pan A C, Arlow D H, Borhani D W, Maragakis P, Shan Y, Xu H, Shaw D E 2011 Proc. Natl. Acad. Sci. USA 108 13118
 Google Scholar
Google Scholar
[27] Gu Y, Shrivastava I H, Amara S G, Bahar I 2009 Proc. Natl. Acad. Sci. USA 106 2589
 Google Scholar
Google Scholar
[28] Latorraca N R, Fastman N M, Venkatakrishnan A J, Frommer W B, Dror R O, Feng L 2017 Cell 169 96
 Google Scholar
Google Scholar
[29] Stelzl L S, Fowler P W, Sansom M S, Beckstein O 2014 J. Mol. Biol. 426 735
 Google Scholar
Google Scholar
[30] Buch I, Giorgino T, De Fabritiis G 2011 Proc. Natl. Acad. Sci. USA 108 10184
 Google Scholar
Google Scholar
[31] Liang R, Swanson J M J, Madsen J J, Hong M, DeGrado W F, Voth G A 2016 Proc. Natl. Acad. Sci. USA 113 E6955
 Google Scholar
Google Scholar
[32] Suomivuori C M, Gamiz-Hernandez A P, Sundholm D, Kaila V R I 2017 Proc. Natl. Acad. Sci. USA 114 7043
 Google Scholar
Google Scholar
[33] Tajkhorshid E, Nollert P, Jensen M Ø, Miercke L J W, O’Connell J, Stroud R M, Schulten K 2002 Science 296 525
 Google Scholar
Google Scholar
[34] Watanabe A, Choe S, Chaptal V, Rosenberg J M, Wright E M, Grabe M, Abramson J 2010 Nature 468 988
 Google Scholar
Google Scholar
[35] Dedmon M M, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson C M 2005 J. Am. Chem. Soc. 127 476
 Google Scholar
Google Scholar
[36] Nguyen H D, Hall C K 2004 Proc. Natl. Acad. Sci. USA 101 16180
 Google Scholar
Google Scholar
[37] Levitt M 1983 J. Mol. Biol. 168 595
 Google Scholar
Google Scholar
[38] Sugita Y, Okamoto Y 1999 Chem. Phys. Lett. 314 141
 Google Scholar
Google Scholar
[39] Rhee Y M, Pande V S 2003 Biophys. J. 84 775
 Google Scholar
Google Scholar
[40] Zhang W, Wu C, Duan Y 2005 J. Chem. Phys. 123 154105
 Google Scholar
Google Scholar
[41] Zhou R 2006 Protein Folding Protocols (Humana Totowa, NJ: Springer) pp205−223
[42] Sindhikara D, Meng Y, Roitberg A E 2008 J. Chem. Phys. 128 024103
 Google Scholar
Google Scholar
[43] Buchete N V, Hummer G 2008 Phys. Rev. E 77 030902
 Google Scholar
Google Scholar
[44] Rosta E, Hummer G 2009 J. Chem. Phys. 131 165102
 Google Scholar
Google Scholar
[45] Stelzl L S, Hummer G 2017 J. Chem. Theory Comput. 13 3927
 Google Scholar
Google Scholar
[46] Yang L J, Gao Q Y 2009 J. Chem. Phys. 131 214109
 Google Scholar
Google Scholar
[47] Yang L, Liu C W, Shao Q, Zhang J, Gao Y Q 2015 Acc. Chem. Res. 48 947
 Google Scholar
Google Scholar
[48] Yang Y I, Zhang J, Che X, Yang L J, Gao Y Q 2016 J. Chem. Phys. 144 094105
 Google Scholar
Google Scholar
[49] Yang Y I, Shao Q, Zhang J, Yang L J, Gao Y Q 2019 J. Chem. Phys. 151 070902
 Google Scholar
Google Scholar
[50] Huber T, Torda A E, Van Gunsteren W F 1994 J. Comput. -Aided Mol. Des. 8 695
 Google Scholar
Google Scholar
[51] Wada T, Kuroda K, Yoshida Y, Ogasawara K, Ogawa A, Endo S 2006 Neurosurg. Rev. 29 242
 Google Scholar
Google Scholar
[52] Hansen H S, Hünenberger P H 2010 J. Comput. Chem. 31 1
 Google Scholar
Google Scholar
[53] Perić-Hassler L, Hansen H S, Baron R, Hünenberger P H 2010 Carbohydr. Res. 345 1781
 Google Scholar
Google Scholar
[54] Grubmüller H 1995 Phys. Rev. E 52 2893
 Google Scholar
Google Scholar
[55] Schulze B G, Grubmüller H, Evanseck J D 2000 J. Am. Chem. Soc. 122 8700
 Google Scholar
Google Scholar
[56] Bouvier B, Grubmüller H 2007 Biophys. J. 93 770
 Google Scholar
Google Scholar
[57] Barducci A, Bonomi M, Parrinello M 2011 WIREs Comput. Mol. Sci. 1 826
 Google Scholar
Google Scholar
[58] Tiwary P, Parrinello M 2013 Phys. Rev. Lett. 111 230602
 Google Scholar
Google Scholar
[59] Bussi G, Laio A 2020 Nat. Rev. Phys. 2 200
 Google Scholar
Google Scholar
[60] Miao Y, Feher V A, McCammon J A 2015 J. Chem. Theory Comput. 11 3584
 Google Scholar
Google Scholar
[61] Miao Y, McCammon J A 2017 Annu. Rep. Comput. Chem. 13 231
 Google Scholar
Google Scholar
[62] Wang J, Arantes P R, Bhattarai A, et al. 2021 Wiley Interdiscip. Rev.: Comput. Mol. Sci. 11 e1521
 Google Scholar
Google Scholar
[63] Naritomi Y, Sotaro F 2011 J. Chem. Phys. 134 065101
 Google Scholar
Google Scholar
[64] Schwantes C R, Vijay S P 2013 J. Chem. Theory Comput. 9 2000
 Google Scholar
Google Scholar
[65] Perez-Hernandez G, Paul F, Giorgino T, Fabritiis G D, Noé F 2013 J. Chem. Phys. 139 015102
 Google Scholar
Google Scholar
[66] Bowman G R, Huang X H, Pande V S 2009 Methods 49 197
 Google Scholar
Google Scholar
[67] Metzner P, Noé F, Schütte C 2009 Phys. Rev. E 80 021106
 Google Scholar
Google Scholar
[68] Pande V S, Beauchamp K A, Bowman G R 2010 Methods 52 99
 Google Scholar
Google Scholar
[69] Prinz J H, Wu H, Sarich M, Keller B, Senne M, Held M, Chodera J D, Schütte C, Noé F 2011 J. Chem. Phys. 134 174105
 Google Scholar
Google Scholar
[70] Kellogg E H, Lange O F, Baker D 2012 J. Phys. Chem. B 116 11405
 Google Scholar
Google Scholar
[71] Yao Y, Cui R Z, Bowman G R, Silva D A, Sun J, Huang X H 2013 J. Chem. Phys. 138 174106
 Google Scholar
Google Scholar
[72] McGibbon R T, Schwantes C R, Pande V S 2014 J. Phys. Chem. B 118 6475
 Google Scholar
Google Scholar
[73] Nuske F, Keller B G, Pérez-Hernández G, Mey A S J, Noé F 2014 J. Chem. Theory Comput. 10 1739
 Google Scholar
Google Scholar
[74] Sheong F K, Silva D A, Meng L, Zhao Y, Huang X H 2015 J. Chem. Theory Comput. 11 17
 Google Scholar
Google Scholar
[75] Zhu L Z, Sheong F K, Zeng X, Huang X H 2016 Phys. Chem. Chem. Phys. 18 30228
 Google Scholar
Google Scholar
[76] Wang W, Cao S, Zhu L Z, Huang X H 2018 WIREs Comput. Mol. Sci. 8 e1343
 Google Scholar
Google Scholar
[77] Husic B E, Pande V S 2018 J. Am. Chem. Soc. 140 2386
 Google Scholar
Google Scholar
[78] Konovalov K A, Unarta I C, Cao S, Goonetilleke E C, Huang X H 2021 J. Am. Chem. Soc. Au. 1 1330
 Google Scholar
Google Scholar
[79] E W, Zhou X 2011 Nonlinearity 24 1831
 Google Scholar
Google Scholar
[80] Samanta A, Chen M, Yu T Q, Tuckerman M E 2014 J. Chem. Phys. 140 164109
 Google Scholar
Google Scholar
[81] Chen M, Yu T Q, Tuckerman M E 2015 Proc. Natl. Acad. Sci. USA 112 3235
 Google Scholar
Google Scholar
[82] E W, Ren W, Vanden-Eijnden E 2002 Phys. Rev. B 66 052301
 Google Scholar
Google Scholar
[83] E W, Ren W, Vanden-Eijnden E 2005 J. Phys. Chem. B 109 6688
 Google Scholar
Google Scholar
[84] Maragliano L, Fischer A, Vanden-Eijnden E, Ciccotti G 2006 J. Chem. Phys. 125 024106
 Google Scholar
Google Scholar
[85] Ren W, Vanden-Eijnden E 2007 J. Chem. Phys. 126 164103
 Google Scholar
Google Scholar
[86] Maragliano L, Vanden-Eijnden E 2007 Chem. Phys. Lett. 446 182
 Google Scholar
Google Scholar
[87] Pan A C, Sezer D, Roux B 2008 J. Phys. Chem. B 112 3432
 Google Scholar
Google Scholar
[88] Chen C, Huang Y, Xiao Y 2012 Phys. Rev. E 86 031901
 Google Scholar
Google Scholar
[89] Chen C, Huang Y, Ji X, Xiao Y 2013 J. Chem. Phys. 138 164122
 Google Scholar
Google Scholar
[90] Chen C J, Huang Y Z, Jiang X W, Xiao Y 2014 J. Chem. Phys. 141 154109
 Google Scholar
Google Scholar
[91] Zhu L Z, Sheong F K, Cao S, Liu S, Unarta I C, Huang X H 2019 J. Chem. Phys. 150 124105
 Google Scholar
Google Scholar
[92] Xi K, Hu Z, Wu Q, Wei M, Qian R, Zhu L Z 2021 J. Chem. Theory Comput. 17 5301
 Google Scholar
Google Scholar
[93] Wang L, Xi K, Zhu L Z, Da L T 2022 J. Chem. Inf. Model. 62 3213
 Google Scholar
Google Scholar
[94] Xi K, Zhu L Z 2022 Int. J. Mol. Sci. 23 14628
 Google Scholar
Google Scholar
[95] Xi K, Zhu L Z 2023 A Practical Guide to Recent Advances in Multiscale Modeling and Simulation of Biomolecules (AIP Publishing) pp9-1−9-24
[96] Vanden-Eijnden E 2006 Computer Simulations in Condensed Matter: From Materials to Chemical Biology (Berlin: Springer) pp453−493
[97] Vanden-Eijnden E 2010 Annu. Rev. Phys. Chem. 61 391
 Google Scholar
Google Scholar
[98] Dellago C, Bolhuis P G, Csajka F S, Chandler D 1998 J. Chem. Phys. 108 1964
 Google Scholar
Google Scholar
[99] Bolhuis P G, Dellago C, Chandler D 1998 Faraday Discuss. 110 421
 Google Scholar
Google Scholar
[100] Dellago C, Bolhuis P G, Chandler D 1999 J. Chem. Phys. 110 6617
 Google Scholar
Google Scholar
[101] Dellago C, Bolhuis P G, Geissler P L 2002 Adv. Chem. Phys. 123 1
 Google Scholar
Google Scholar
[102] Noé F, Tkatchenko A, Müller K R, Clementi C 2020 Annu. Rev. Phys. Chem. 71 361
 Google Scholar
Google Scholar
[103] AlQuraishi M, Sorger P K 2021 Nat. Methods 18 1169
 Google Scholar
Google Scholar
[104] Karniadakis G E, Kevrekidis I G, Lu L, Perdikaris P, Wang S, Yang L 2021 Nat. Rev. Phys. 3 422
 Google Scholar
Google Scholar
[105] Ju F, Zhu J, Shao B, Kong L, Liu T Y, Zheng W M, Bu D 2021 Nat. Commum. 12 2535
 Google Scholar
Google Scholar
[106] Huang B, Xu Y, Hu X H, Liu Y R, Liao S H, Zhang J H, Huang C D, Hong J J, Chen Q, Liu H Y 2022 Nature 602 523
 Google Scholar
Google Scholar
[107] Du Z, Su H, Wang W, Ye L, Wei H, Peng Z, Anishchenko I, Baker D, Yang J 2021 Nat. Prot. 16 5634
 Google Scholar
Google Scholar
[108] Dai M, Dong Z, Xu K, Zhang Q C 2023 J. Mol. Biol. 435 168059
 Google Scholar
Google Scholar
[109] Yuan Q M, Chen J W, Zhao H Y, Zhou Y Q, Yang Y D 2022 Bioinformatics 38 125
 Google Scholar
Google Scholar
[110] Su M Y, Feng G, Liu Z, Li Y, Wang R 2020 J. Chem. Inf. Model. 60 1122
 Google Scholar
Google Scholar
[111] Zeng C, Jian Y, Vosoughi S, Zeng C, Zhao Y 2023 Nat. Commum. 14 1060
 Google Scholar
Google Scholar
[112] Noé F, Olsson S, Köhler J, Wu H 2019 Science 365 6457
 Google Scholar
Google Scholar
[113] Jung H, Covino R, Arjun A, Leitold C, Dellago C, Bolhuis P G, Hummer G 2023 Nat. Comput. Sci. 3 334
 Google Scholar
Google Scholar
[114] Weiss D R, Levitt M 2009 J. Mol. Biol. 385 665
 Google Scholar
Google Scholar
[115] Isralewitz B, Gao M, Schulten K 2001 Curr. Opin. Struct. Biol. 11 224
 Google Scholar
Google Scholar
[116] Schlitter J, Engels M, Krüger P 1994 J. Mol. Graph. 12 84
 Google Scholar
Google Scholar
[117] Torrie G M, Valleau J P 1977 J. Comput. Phys. 23 187
 Google Scholar
Google Scholar
[118] Ryckaert J P, Ciccotti G, Berendsen H J 1977 J. Comput. Phys. 23 327
 Google Scholar
Google Scholar
[119] Babin V, Roland C, Sagui C 2008 J. Chem. Phys. 128 134101
 Google Scholar
Google Scholar
[120] Branduardi D, Gervasio F L, Parrinello M 2007 J. Chem. Phys. 126 054103
 Google Scholar
Google Scholar
[121] Applegate D L, Bixby R E, Chvátal V, Cook W J 2011 The Traveling Salesman Problem: A Computational Study (Princeton University Press) pp1−58
[122] Cox M A A, Cox T F 2008 Handbook of Data Visualization (Berlin: Springer) pp315−347
[123] Barducci A, Bussi G, Parrinello M 2008 Phys. Rev. Lett. 100 020603
 Google Scholar
Google Scholar
[124] Fischmann T O, Smith C K, Mayhood T W, Myers J E, Reichert J P, Mannarino A, Carr D, Zhu H, Wong J, Yang R S, Le H V, Madison V S 2009 Biochemistry 48 2661
 Google Scholar
Google Scholar
[125] Hanrahan A J, Sylvester B E, Chang M T, et al. 2020 Cancer Res. 80 4233
 Google Scholar
Google Scholar
[126] Schmidt M, Lipson H 2009 Science 324 81
 Google Scholar
Google Scholar
[127] Jung H, Okazaki K, Hummer G 2017 J. Chem. Phys. 147 152716
 Google Scholar
Google Scholar
[128] Swenson D W H, Prinz J H, Noe F, Chodera J D, Bolhuis P G 2019 J. Chem. Theory Comput. 15 813
 Google Scholar
Google Scholar
[129] Swenson D W H, Prinz J H, Noe F, Chodera J D, Bolhuis P G 2019 J. Chem. Theory Comput. 15 837
 Google Scholar
Google Scholar
[130] Glielmo A, Husic B E, Rodriguez A, Clementi C, Noé F, Laio A 2021 Chem. Rev. 121 9722
 Google Scholar
Google Scholar
计量
- 文章访问数: 8083
- PDF下载量: 249
- 被引次数: 0














 下载:
下载: