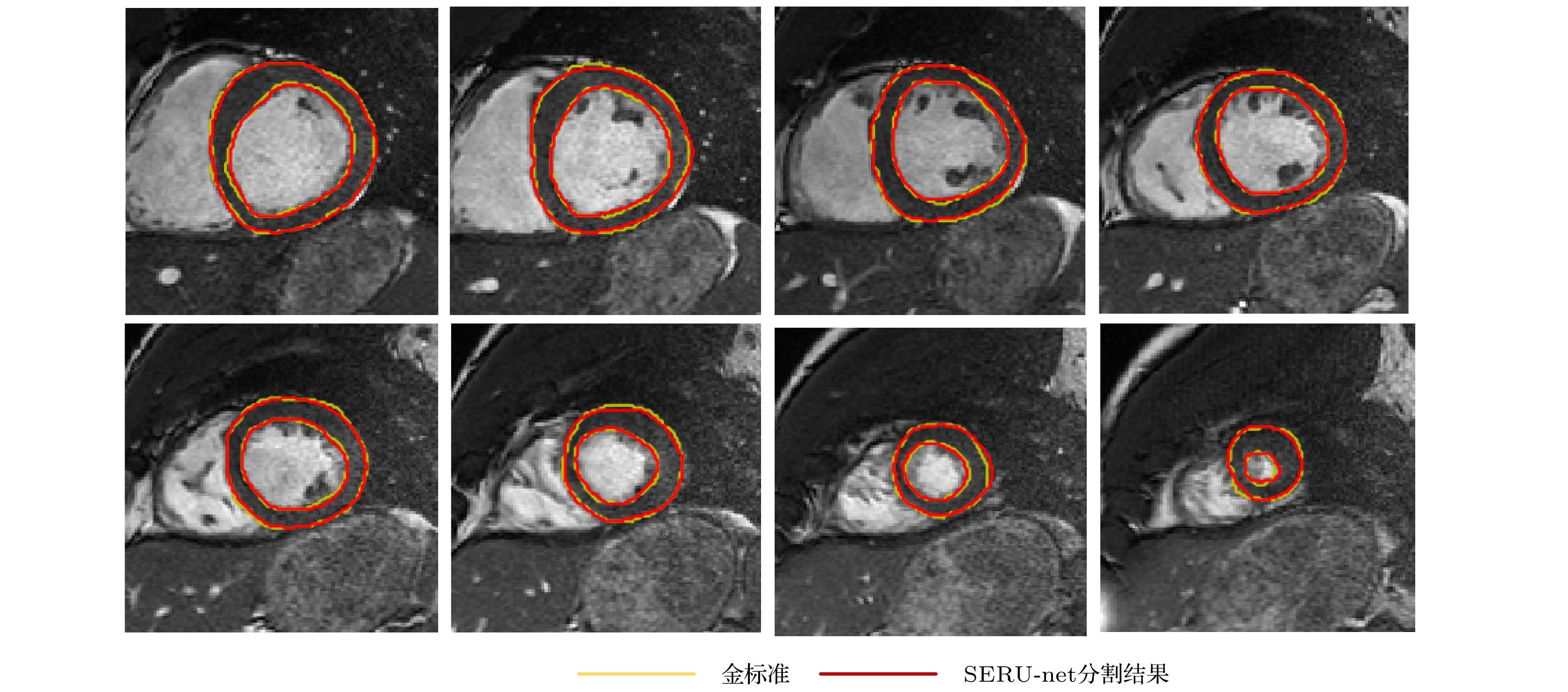
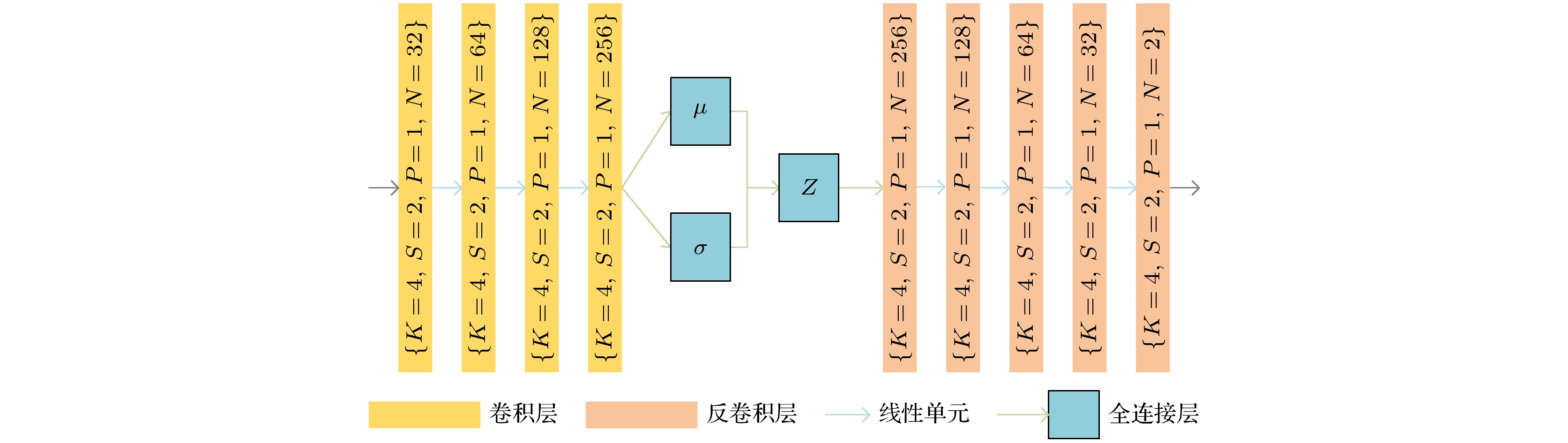
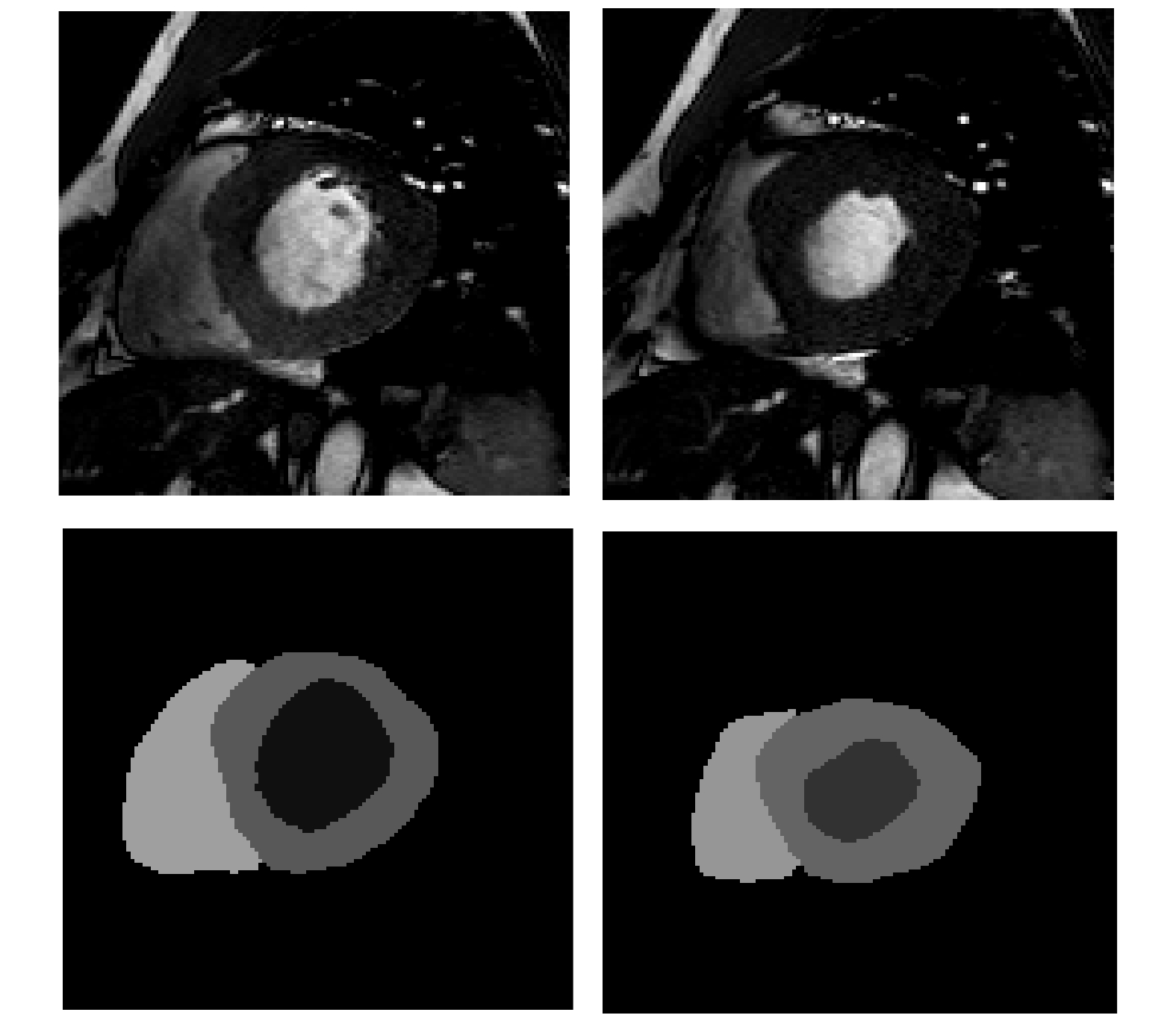
-
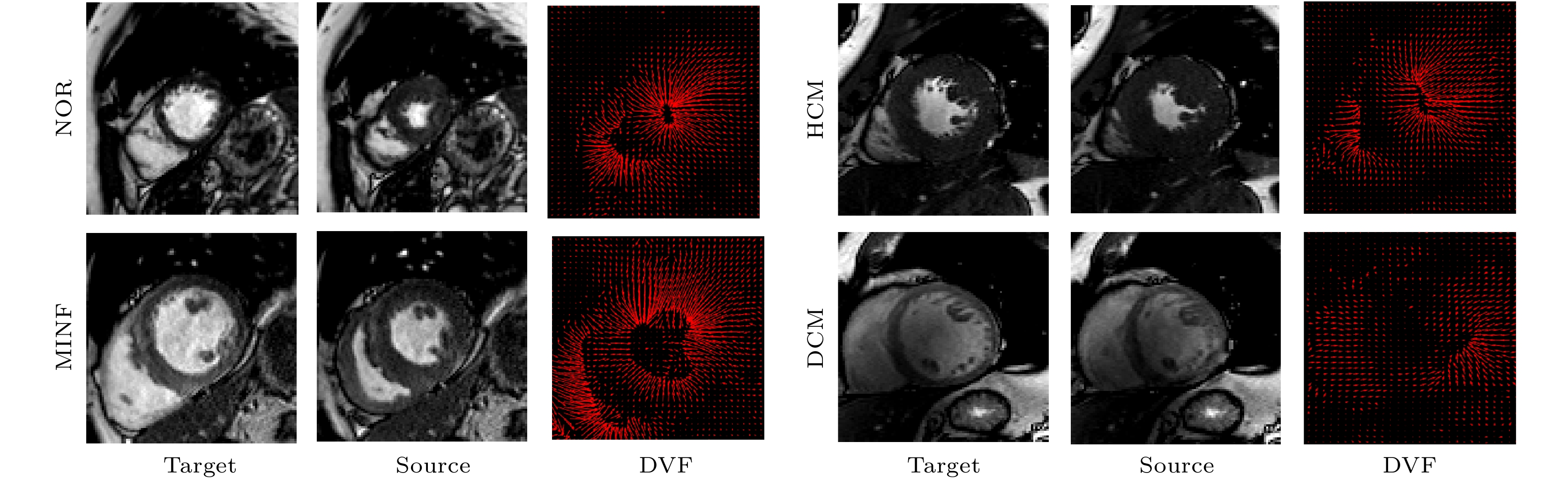
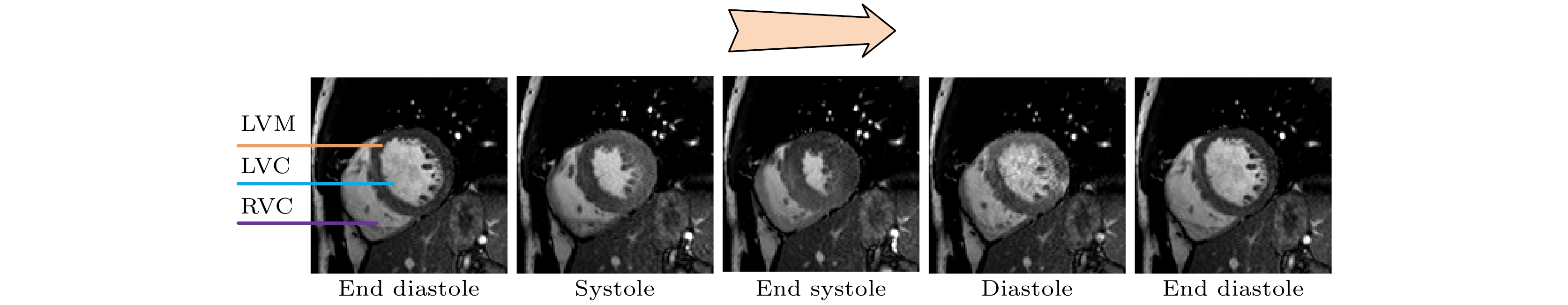
心血管疾病(cardiovascular diseases, CVDs)的高发病率和高死亡率已经严重影响了人类的生存质量. 如何评估心脏功能、辅助临床CVDs诊疗和预后评估, 是一个迫切需要解决的问题 . 针对这个问题, 本文在前期心脏电影磁共振(cardiac cine magnetic resonance, CCMR)图像左心肌分割的基础上, 提出一种基于位移流U-Net(DispFlow_UNet)和生物力学变分自动编码器(variational autoencoder, VAE)的左心肌运动追踪方法: DispFlow_UNet_VAE. 主要研究内容有: 1) 搭建压缩激励残差U-net网络精准分割左心肌, 根据分割结果计算心室体积、心肌质量等, 评估心脏整体功能; 2) 根据DispFlow_UNet_VAE估计CCMR图像连续帧之间的左心室运动, 结合左心肌分割掩膜得到左心肌密集位移场; 3)利用模拟数据真实位移场、临床数据集对追踪结果进行对比和评估. 结果表明, 本文追踪算法具有较高的精度和泛化能力.The high morbidity and mortality of cardiovascular diseases (CVDs) seriously affects the quality of human life. How to asses cardiac function, assist in the diagnosis and treatment of clinical CVDs and evaluate prognosis is a problem to be solved urgently. In response to this issue, based on previous work of Cardiac Cine Magnetic Resonance (CCMR) image segmentation of the left myocardium (LVM), a robust and accurate LVM motion tracking method (DispFlow_UNet_Flow) with using the displacement flow UNet (DispFlow_UNet) and biomechanics-informed variational autoencoder (VAE) is proposed in this paper. The following are the main research contents: (1) building a compressed excitation residual U-net network to accurately segment LVM, calculating the ventricular volume and myocardial mass according to the segmentation results, and then evaluating the overall cardiac function; (2) reconstructing the dense displacement field (DDF) based on the proposed motion tracking method, and obtaining the LVM dense displacement field by combining the LVM segmentation mask; (3) contrasting and evaluating the motion tracking results by using the true displacement vector field of simulated data and clinical data sets. All the results show that the tracking algorithm proposed in this paper has high precision and generalization capability.
-
Keywords:
- left myocardium tracking /
- deep learning /
- displacement flow U-Net network /
- variational autoencoder
[1] World Health Organization, http://origin.who.int/mediacentrse/factsheets/fs317/en/ [2019−4−17]
[2] 胡盛寿, 高润霖, 刘力生, 朱曼璐, 王文, 王拥军, 吴兆苏, 李惠君, 顾东风, 杨跃进, 郑哲, 陈伟伟 2019 中国循环学杂志 34 209
Hu S S, Gao R L, Liu L S, Zhu M L, Wang W, Wang Y J, Wu Z S, Li H J, Gu D F, Yang Y J, Zheng Z, Chen W W 2019 Chin. Circ. J. 34 209
[3] Stathogiannis K, Mor-Avi V, Rashedi N, Lang R M, Patel A R 2020 Med. Image Anal. 68 190
[4] Peng P, Lekadir K, Goova A, Shao L, Petersen S E, Frangi A F 2016 Magn. Reson. Mater. Phys. , Biol. Med. 29 155
[5] Frangi A F, Niessen W J, Viergever M A 2001 IEEE Trans. Med. Imaging 20 2
 Google Scholar
Google Scholar
[6] Young, Alistair A 2006 Curr. Cardiol. Rev. 2 271
 Google Scholar
Google Scholar
[7] Underwood S R, Rees R S, Savage P E, Klipstein R H, Firmin D N, Fox K M, Poole-Wilson P A, Longmore D B 1986 Br. Heart J. 56 334
 Google Scholar
Google Scholar
[8] Darasz K H, Underwood S R, Bayliss J, Forbat S M, Keegan J, Poole-Wilson P A, Sutton G C 2002 Int. J. Cardiovas. Imaging 18 135
 Google Scholar
Google Scholar
[9] Castillo E, Lima J, Bluemke D A 2003 Radiographics 23 S127
 Google Scholar
Google Scholar
[10] Mcveigh E R, Zerhouni E A 1991 Radiol. 180 677
 Google Scholar
Google Scholar
[11] Wang H, Amini A A 2012 IEEE Trans. Med. Imaging 31 487
 Google Scholar
Google Scholar
[12] Yu H, Sun S, Yu H, Chen X, Shi H, Huang T, Chen T 2020 arXiv: 2003.04492 v2 [cs. CV]
[13] Afshin M, Ben Ayed I, Punithakumar K, Law M, Islam A, Goela A, Peters T, Li S 2014 IEEE Trans. Med. Imaging 33 481
 Google Scholar
Google Scholar
[14] Wang L, Clarysse P, Liu Z, Gao B, Delachartre P 2019 Med. Image Anal. 57 136
 Google Scholar
Google Scholar
[15] Yousefi-Banaem H, Kermani S, Asiaei S, Sanei H 2017 Comput. Biol. Med. 80 56
 Google Scholar
Google Scholar
[16] Tobon-Gomez C, Craene M D, Mcleod K, Tautz L, Shi W, Hennemuth A, Prakosa A, Wang H, Carr-White G, Kapetenakis S, Muller-Lutz A, Rasche V, Friman O, Mansi T, Sermesant M, Zhuang X, Ourselin S, Peitgen H, Pennec X, Razavi R, Ruecjert D, Frangi A F, Rhode K 2013 Med. Image Anal. 17 632
 Google Scholar
Google Scholar
[17] Puyol-Anton E, Ruijsink B, Bai W, Langet H, Sinclair M, De-Craene M, Schnabel J, Piro P, King A 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) Washington, USA, April 1, 2018 p1139
[18] Mcleod K, Sermesant M, Beerbaum P, Pennec X 2015 IEEE Trans. Med. Imaging 34 1562
 Google Scholar
Google Scholar
[19] Qin C, Bai W, Schlemper J, Petersen S, Piechnik S, Neubauer S, Rueckert D 2018 Medical Image Computing and Computer Assisted Intervention-MICCAI 2018 Granada, Spain, September 16, 2018 p472
[20] Zheng Q, Delingette H, Ayache N 2019 Med. Image Anal. 56 80
 Google Scholar
Google Scholar
[21] Vos B D, Berendsen F F, Viergever M A , Staring M, Igum I 2017 ML-CDS 2017: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support Québec City, Canada, September 10, 2017 p204
[22] Qiao M, Wang Y, Guo Y, Huang L, Xia L, Tan Q 2020 Med. Phys. 47 4189
 Google Scholar
Google Scholar
[23] Chen P, Chen X, Chen E, Yu H, Chen T, Sun S 2020 arXiv: 2008.07579v1 [eess. IV]
[24] Ronneberger O, Fischer P, Brox T 2015 Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015 Munich, Germany October 5–9, 2015 p234
[25] 王慧 2020 硕士学位论文 (上海: 上海理工大学)
Wang H 2020 M. S. Thesis (Shanghai: University of Shanghai for Science and Technology) (in Chinese)
[26] Qiu H, Qin C, Folgoc L L, Hou B, Schlemper Jo, Ruechert D 2019 STACOM 2019: Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges Shenzhen, China, October 13, 2019 p186
[27] Krebs J, Delingette H E, Mailhe B, Ayache N, Mansi T 2019 IEEE Trans. Med. Imaging 38 2165
 Google Scholar
Google Scholar
[28] Kingma D P, Welling M 2014 2nd International Conference on Learning Representations, ICLR 2014-Conference Track Proceedings Banff, Canada, April 14–16, 2014
[29] Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng, Pheng-Ann, Cetin I, Lekadir K, Camara O, Ballester M 2018 IEEE Trans. Med. Imaging 37 2514
 Google Scholar
Google Scholar
[30] Duchateau N, Sermesant M, Delingette H, Ayache N 2017 IEEE Trans. Med. Imaging 37 755
 Google Scholar
Google Scholar
[31] Rueckert D, Sonoda L I, Hayes C, Hill D L G, Leach M O, Hawkes D J 1999 IEEE Trans. Med. Imaging 18 712
 Google Scholar
Google Scholar
-
表 1 实验数据
Table 1. The experimental data.
数据集 数据量 相位数 层数 磁共振扫描仪 ACDC 100 12—35 9—10 1.5 T Siemens Area
3.0 T Siemens
Trio Tim临床数据 75 20—28 6—10 1.5 T GE 合成MRI数据 15 30 9—14 无 表 2 ACDC数据集分配情况
Table 2. ACDC data set allocation.
第n重交叉验证(Foldn) Fold1 Fold2 Fold3 Fold4 Fold5 内部数据data_ int HCM MINFNOR RVA DCM MINFNOR RVA DCM HCMNOR RVA DCM HCMMINF RVA DCM HCMMINF NOR 外部数据 data_ ext DCM HCM MINF NOR RVA 训练集 data_int×0.8 验证集 data_int×0.1 预测集 data_int×0.1 + data_ext×1 表 3 左心室功能指参数 (均值±标准差)
Table 3. Left ventricular function parameters (Mean ± standard deviation).
EDV
/mLESV/mL SV
/mLEF
/%ED_LVM/L·min–1 ES_LVM
/g97.94
±34.7338.133
±21.3061.39
±24.4362.90
±15.388.23
±35.2384.13±33.22 表 4 不同追踪方法Dice系数、MCD和HD的对比
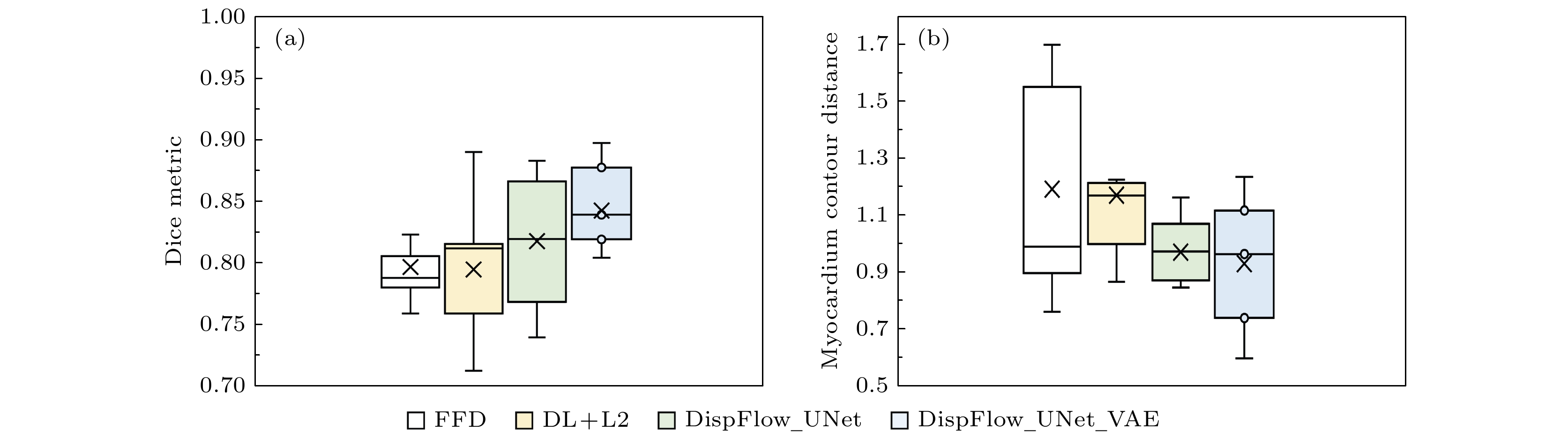
Table 4. Comparison of Dice coefficients, MCD and HD of different tracking methods.
方法 Dice MCD HD LVC LVM LVC LVM LVC LVM FFD 0.920 (0.029) 0.797 (0.034) 1.256 (0.387) 1.192 (0.392) 3.431 (0.688) 3.439 (1.181) DL+L2 0.912 (0.037) 0.800 (0.057) 1.340 (0.428) 1.171 (0.286) 3.699 (1.099) 3.285 (0.717) DispFlow_UNet 0.925 (0.024) 0.818 (0.054) 1.203 (0.329) 0.971 (0.120) 3.347 (0.785) 2.906 (0.358) DispFlow_UNet_VAE 0.946 (0.016) 0.843 (0.031) 0.924 (0.279) 0.931 (0.239) 2.991 (0.960) 3.178 (0.744) 表 5 临床数据集上的模型泛化性能
Table 5. Model generalization performance on clinical datasets.
数据集 Dice MCD LVC LVM RVC LVC LVM RVC ACDC 0.946 (0.016) 0.843 (0.031) 0.876 (0.078) 0.924 (0.279) 0.931 (0.239) 1.348 (0.858) 临床数据 0.882 (0.022) 0.836 (0.059) 0.850 (0.093) 1.874 (0.383) 1.079 (0.262) 1.427 (0.579) -
[1] World Health Organization, http://origin.who.int/mediacentrse/factsheets/fs317/en/ [2019−4−17]
[2] 胡盛寿, 高润霖, 刘力生, 朱曼璐, 王文, 王拥军, 吴兆苏, 李惠君, 顾东风, 杨跃进, 郑哲, 陈伟伟 2019 中国循环学杂志 34 209
Hu S S, Gao R L, Liu L S, Zhu M L, Wang W, Wang Y J, Wu Z S, Li H J, Gu D F, Yang Y J, Zheng Z, Chen W W 2019 Chin. Circ. J. 34 209
[3] Stathogiannis K, Mor-Avi V, Rashedi N, Lang R M, Patel A R 2020 Med. Image Anal. 68 190
[4] Peng P, Lekadir K, Goova A, Shao L, Petersen S E, Frangi A F 2016 Magn. Reson. Mater. Phys. , Biol. Med. 29 155
[5] Frangi A F, Niessen W J, Viergever M A 2001 IEEE Trans. Med. Imaging 20 2
 Google Scholar
Google Scholar
[6] Young, Alistair A 2006 Curr. Cardiol. Rev. 2 271
 Google Scholar
Google Scholar
[7] Underwood S R, Rees R S, Savage P E, Klipstein R H, Firmin D N, Fox K M, Poole-Wilson P A, Longmore D B 1986 Br. Heart J. 56 334
 Google Scholar
Google Scholar
[8] Darasz K H, Underwood S R, Bayliss J, Forbat S M, Keegan J, Poole-Wilson P A, Sutton G C 2002 Int. J. Cardiovas. Imaging 18 135
 Google Scholar
Google Scholar
[9] Castillo E, Lima J, Bluemke D A 2003 Radiographics 23 S127
 Google Scholar
Google Scholar
[10] Mcveigh E R, Zerhouni E A 1991 Radiol. 180 677
 Google Scholar
Google Scholar
[11] Wang H, Amini A A 2012 IEEE Trans. Med. Imaging 31 487
 Google Scholar
Google Scholar
[12] Yu H, Sun S, Yu H, Chen X, Shi H, Huang T, Chen T 2020 arXiv: 2003.04492 v2 [cs. CV]
[13] Afshin M, Ben Ayed I, Punithakumar K, Law M, Islam A, Goela A, Peters T, Li S 2014 IEEE Trans. Med. Imaging 33 481
 Google Scholar
Google Scholar
[14] Wang L, Clarysse P, Liu Z, Gao B, Delachartre P 2019 Med. Image Anal. 57 136
 Google Scholar
Google Scholar
[15] Yousefi-Banaem H, Kermani S, Asiaei S, Sanei H 2017 Comput. Biol. Med. 80 56
 Google Scholar
Google Scholar
[16] Tobon-Gomez C, Craene M D, Mcleod K, Tautz L, Shi W, Hennemuth A, Prakosa A, Wang H, Carr-White G, Kapetenakis S, Muller-Lutz A, Rasche V, Friman O, Mansi T, Sermesant M, Zhuang X, Ourselin S, Peitgen H, Pennec X, Razavi R, Ruecjert D, Frangi A F, Rhode K 2013 Med. Image Anal. 17 632
 Google Scholar
Google Scholar
[17] Puyol-Anton E, Ruijsink B, Bai W, Langet H, Sinclair M, De-Craene M, Schnabel J, Piro P, King A 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) Washington, USA, April 1, 2018 p1139
[18] Mcleod K, Sermesant M, Beerbaum P, Pennec X 2015 IEEE Trans. Med. Imaging 34 1562
 Google Scholar
Google Scholar
[19] Qin C, Bai W, Schlemper J, Petersen S, Piechnik S, Neubauer S, Rueckert D 2018 Medical Image Computing and Computer Assisted Intervention-MICCAI 2018 Granada, Spain, September 16, 2018 p472
[20] Zheng Q, Delingette H, Ayache N 2019 Med. Image Anal. 56 80
 Google Scholar
Google Scholar
[21] Vos B D, Berendsen F F, Viergever M A , Staring M, Igum I 2017 ML-CDS 2017: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support Québec City, Canada, September 10, 2017 p204
[22] Qiao M, Wang Y, Guo Y, Huang L, Xia L, Tan Q 2020 Med. Phys. 47 4189
 Google Scholar
Google Scholar
[23] Chen P, Chen X, Chen E, Yu H, Chen T, Sun S 2020 arXiv: 2008.07579v1 [eess. IV]
[24] Ronneberger O, Fischer P, Brox T 2015 Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015 Munich, Germany October 5–9, 2015 p234
[25] 王慧 2020 硕士学位论文 (上海: 上海理工大学)
Wang H 2020 M. S. Thesis (Shanghai: University of Shanghai for Science and Technology) (in Chinese)
[26] Qiu H, Qin C, Folgoc L L, Hou B, Schlemper Jo, Ruechert D 2019 STACOM 2019: Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges Shenzhen, China, October 13, 2019 p186
[27] Krebs J, Delingette H E, Mailhe B, Ayache N, Mansi T 2019 IEEE Trans. Med. Imaging 38 2165
 Google Scholar
Google Scholar
[28] Kingma D P, Welling M 2014 2nd International Conference on Learning Representations, ICLR 2014-Conference Track Proceedings Banff, Canada, April 14–16, 2014
[29] Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng, Pheng-Ann, Cetin I, Lekadir K, Camara O, Ballester M 2018 IEEE Trans. Med. Imaging 37 2514
 Google Scholar
Google Scholar
[30] Duchateau N, Sermesant M, Delingette H, Ayache N 2017 IEEE Trans. Med. Imaging 37 755
 Google Scholar
Google Scholar
[31] Rueckert D, Sonoda L I, Hayes C, Hill D L G, Leach M O, Hawkes D J 1999 IEEE Trans. Med. Imaging 18 712
 Google Scholar
Google Scholar
计量
- 文章访问数: 6411
- PDF下载量: 75
- 被引次数: 0













 下载:
下载: