-
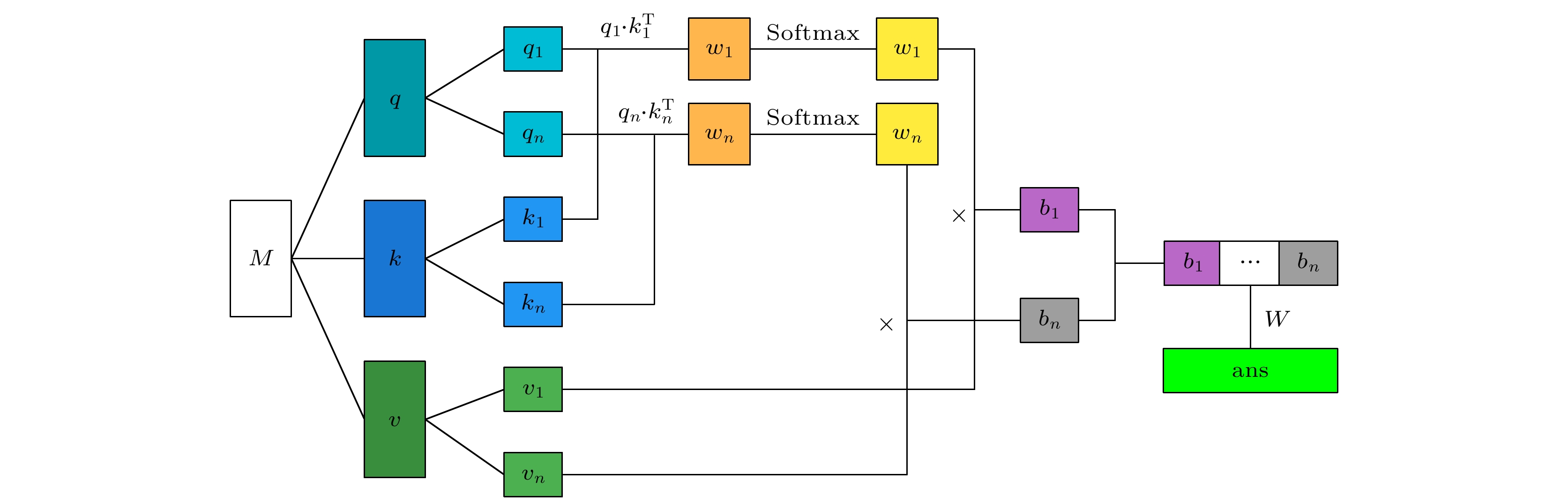
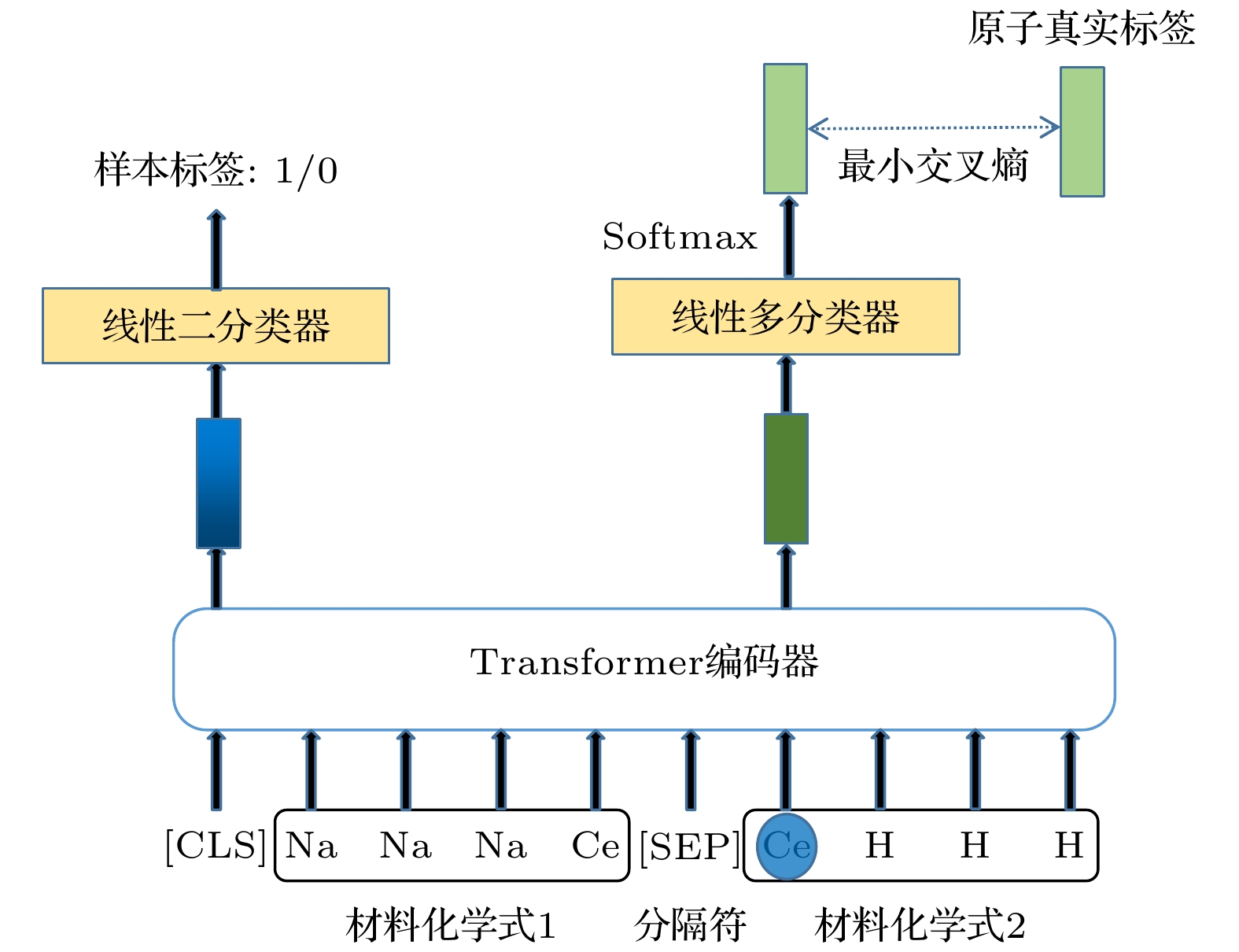
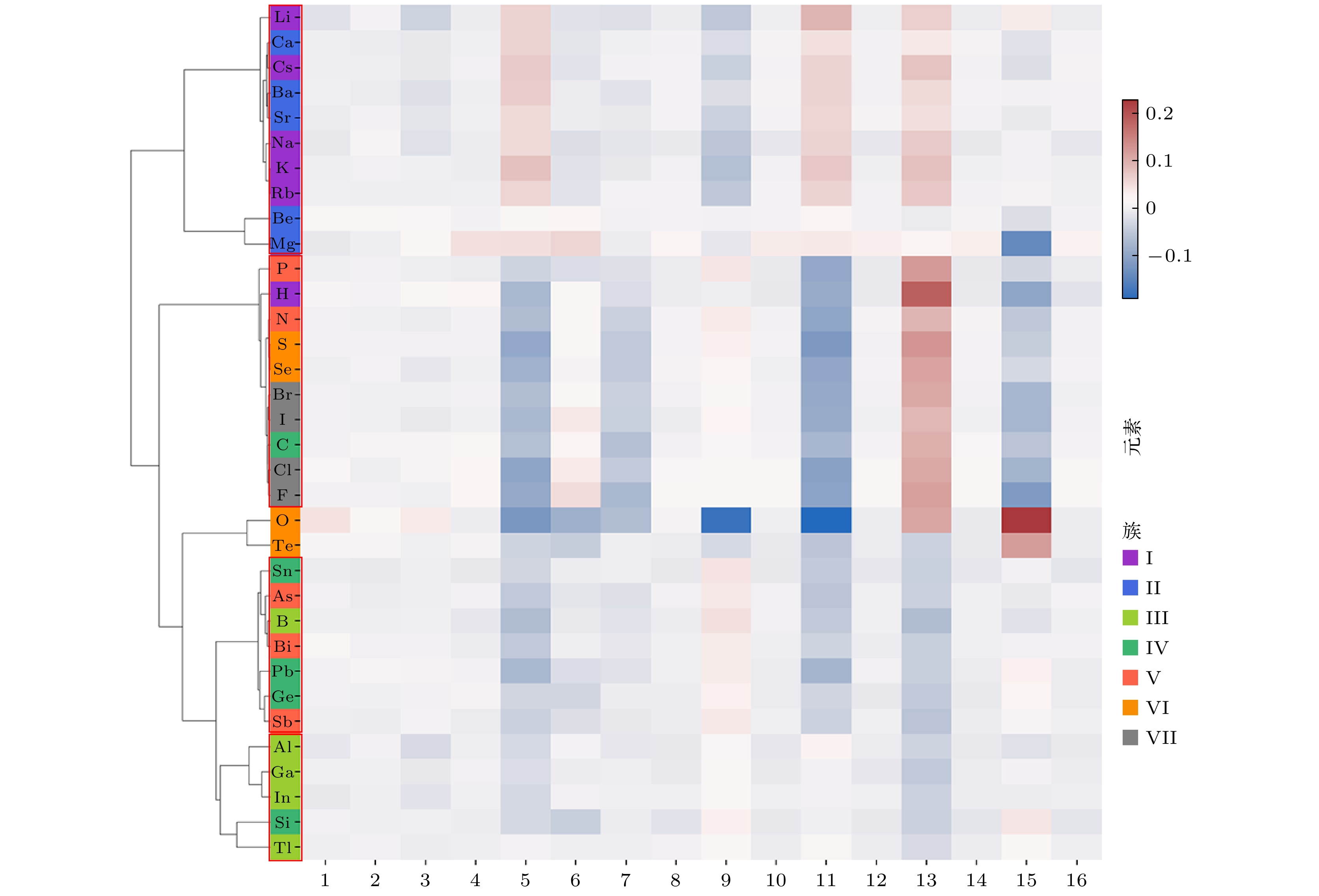
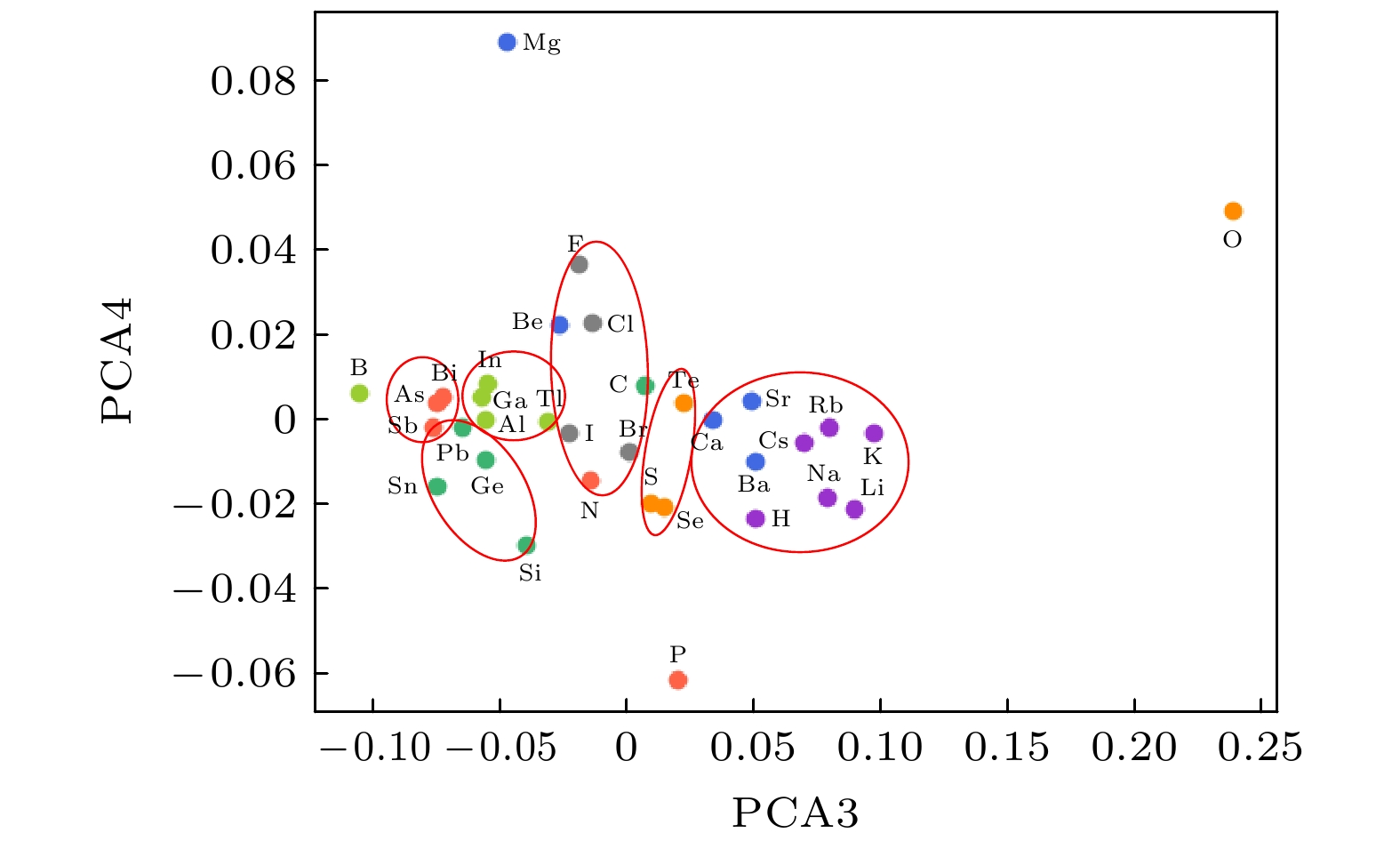
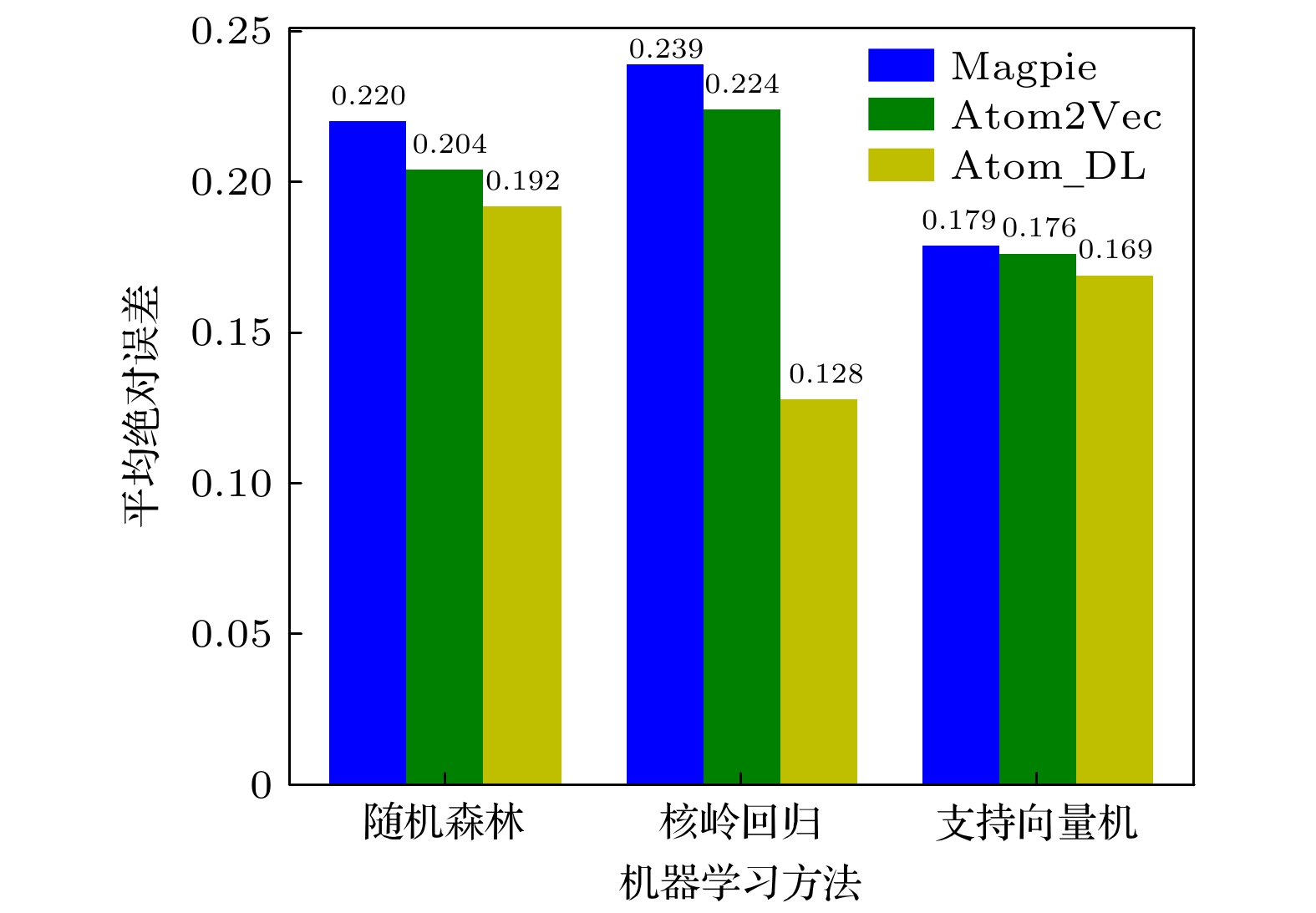
随着人工智能的发展, 机器学习在材料计算中的应用越来越广泛. 将机器学习应用到材料性质预测等任务中首要实现的是获得有效的材料特征表示. 本文采用一种原子特征表示方法, 研究一种低维、密集的分布式原子特征向量, 并用于材料带隙预测任务. 按照材料化学式中原子种类和原子个数, 使用Transformer编码器作为模型结构, 通过训练大量的材料化学式数据, 从而提取参与训练元素的特征. 利用该方法预测Janus结构过渡金属硫族化合物MXY (M代表过渡金属, X, Y是不同硫族元素)二维材料带隙. 基于深度学习得到的原子特征向量比传统的Magpie方法和Atom2Vec方法的预测平均绝对误差更小. 可视化分析和材料性质预测数值实验表明, 本文提出的基于深度学习提取的原子特征表示方法, 可以有效表征材料特征, 并且应用到材料带隙预测任务中.With the development of artificial intelligence, machine learning (ML) is more and more widely used in material computing. To apply ML to the prediction of material properties, the first thing to do is to obtain effective material feature representation. In this paper, an atomic feature representation method is used to study a low-dimensional, densely distributed atomic eigenvector, which is applied to the band gap prediction in material design. According to the types and numbers of atoms in the chemical formula of material, the Transformer Encoder is used as a model structure, and a large number of material chemical formula data are trained to extract the features of the training elements. Through the clustering analysis of the atomic feature vectors of the main group elements, it is found that the element features can be used to distinguish the element categories. The Principal Component Analysis of the atomic eigenvector of the main group element shows that the projection of the atomic eigenvector on the first principal component reflects the outermost electron number corresponding to the element. It illustrates the effectiveness of atomic eigenvector extracted by using the transformer model. Subsequently, the atomic feature representation method is used to represent the material characteristics. Three ML methods named Random Forest (RF), Kernel Ridge Regression (KRR) and Support Vector Regression (SVR) are used to predict the band gap of the two-dimensional transition metal chalcogenide compound MXY (M represents transition metal, X and Y refer to the different chalcogenide elements) with Janus structure. The hyperparameters of ML model are determined by searching for parameters. To obtain stable results, the ML model is tested by 5-fold cross-validation. The results obtained from the three ML models show that the average absolute error of the prediction using atomic feature vectors based on deep learning is smaller than that obtained from the traditional Magpie method and the Atom2Vec method. For the atomic eigenvector method proposed in this paper, the prediction accuracy of the KRR model is better than that of the results obtained from the Magpie method and Atom2Vec method. It shows that the atomic feature vector proposed in this paper has a certain correlation between the features, and is a low-dimensional and densely distributed feature vector. Visual analysis and numerical experiments of material property prediction show that the atomic feature representation method based on deep learning extraction proposed in this paper can effectively characterize the material features and can be applied to the tasks of material band gap prediction.
-
Keywords:
- atomic feature representation /
- deep learning /
- transition metal sulfide /
- band gap
[1] He K M, Zhang X Y, Ren S Q, Sun J 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Las Vegas, NV, USA, June 27–30, 2016 p770
[2] Ren S Q, He K M, Girshick R, Sun J 2017 IEEE Trans. Pattern Anal. Mach. Intell. 39 1137
 Google Scholar
Google Scholar
[3] Devlin J, Chang M W, Lee K, Toutanova K 2019 Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies Minneapolis, USA, June 3–5, 2019 p4171
[4] 郭佳龙, 王宗国, 王彦棡, 赵旭山, 宿彦京, 刘志威 2021 数据与计算发展前沿 3 120
 Google Scholar
Google Scholar
Guo J L, Wang Z G, Wang Y G, Zhao X S, Su Y J, Liu Z W 2021 Frontiers of Data and Computing 3 120
 Google Scholar
Google Scholar
[5] 牛程程, 李少波, 胡建军, 但雅波, 曹卓, 李想 2020 材料导报 34 23100
 Google Scholar
Google Scholar
Niu C C, Li S B, Hu J J, Dan Y B, Cao Z, Li X 2020 Mater. Rep. 34 23100
 Google Scholar
Google Scholar
[6] Hu T T, Song H, Jiang T, Li S B 2020 Symmetry 12 1889
 Google Scholar
Google Scholar
[7] Chen C, Ye W K, Zuo Y X, Zheng C, Ong S P 2019 Chem. Mater. 31 3564
 Google Scholar
Google Scholar
[8] Li S B, Dan Y B, Li X, Hu T T, Dong R Z, Cao Z, Hu J J 2020 Symmetry 12 262
 Google Scholar
Google Scholar
[9] Zhang L F, Han J Q, Wang H, Car R, E W N 2018 Phys. Rev. Lett. 120 143001
 Google Scholar
Google Scholar
[10] de Jong M, Chen W, Notestine R, Persson K, Ceder G, Jain A, Asta M, Gamst A 2016 Sci. Rep. 6 34256
 Google Scholar
Google Scholar
[11] Zhou Q, Tang P Z, Liu S X, Pan J B, Yan Q M, Zhang S C 2018 Proc. Nat1. Acad. Sci. U. S. A. 115 6411
 Google Scholar
Google Scholar
[12] Calfa B A, Kitchin J R 2016 AIChE J. 62 2605
 Google Scholar
Google Scholar
[13] Ward L, Agrawal A, Choudhary A, Wolverton C 2016 NPJ Comput. Mater. 2 16028
 Google Scholar
Google Scholar
[14] Zhuo Y, Mansouri Tehrani A, Brgoch J 2018 J. Phys. Chem. Lett. 9 1668
 Google Scholar
Google Scholar
[15] Hu M X, Yuan J M, Sun T, Huang M, Liang Q Y 2021 Comput. Mater. Sci. 200 110841
 Google Scholar
Google Scholar
[16] Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A N, Kaiser Ł, Polosukhin I 2017 31st Conference on Neural Information Processing Systems (NIPS 2017) Long Beach, CA, USA, December 4–9, 2017 p6000
[17] Saal J E, Kirklin S, Aykol M, Meredig B, Wolverton C 2013 JOM 65 1501
 Google Scholar
Google Scholar
[18] Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z M, Gimelshein N, Antiga L, Desmaison A, Köpf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J J, Chintala S 2019 Proceedings of the 33rd International Conference on Neural Information Processing Systems Vancouver, Canada, December 8–14, 2019 p8026
[19] Wang G, Chernikov A, Glazov M M, Heinz T F, Marie X, Amand T, Urbaszek B 2018 Rev. Mod. Phys. 90 021001
 Google Scholar
Google Scholar
[20] Riis-Jensen A C, Deilmann T, Olsen T, Thygesen K S 2019 ACS Nano 13 13354
 Google Scholar
Google Scholar
[21] Gjerding M N, Taghizadeh A, Rasmussen A, Ali S, Bertoldo F, Deilmann T, Knøsgaard N R, Kruse M, Larsen A H, Manti S, Pedersen T G, Petralanda U, Skovhus T, Svendsen M K, Mortensen J J, Olsen T, Thygesen K S 2021 2D Mater. 8 044002
 Google Scholar
Google Scholar
[22] Haastrup S, Strange M, Pandey M, Deilmann T, Schmidt P S, Hinsche N F, Gjerding M N, Torelli D, Larsen P M, Riis-Jensen A C, Gath J, Jacobsen K W, Jørgen Mortensen J, Olsen T, Thygesen K S 2018 2D Mater. 5 042002
 Google Scholar
Google Scholar
[23] Schütt K T, Glawe H, Brockherde F, Sanna A, Müller K R, Gross E K U 2014 Phys. Rev. B 89 205118
 Google Scholar
Google Scholar
[24] Wu Y R, Li H P, Gan X S 2013 Adv. Mater. Res. 848 122
 Google Scholar
Google Scholar
[25] Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É 2011 J. Mach. Learn. Res. 12 2825
 Google Scholar
Google Scholar
-
表 1 随机森林模型参数
Table 1. The random forest model parameter.
机器学习方法 原子表征方法 随机森林中树的个数 随机森林 Magpie 50 Atom2Vec 80 Atom_DL 10 表 2 核岭回归模型参数
Table 2. Kernel ridge regression model parameter.
机器学习方法 原子表征方法 核函数 多项式核次数 正则化强度 伽马参数 零系数 核岭回归 Magpie 多项式核 1 1 0.010 1.0 Atom2Vec 多项式核 2 1 0.001 0.5 Atom_DL 多项式核 4 1 0.300 1.5 表 3 支持向量机回归模型参数
Table 3. Support vector regression model parameter.
机器学习方法 原子表征方法 核函数 多项式核次数 正则化参数 伽马参数 零系数 支持向量机 Magpie 多项式核 1 0.1 0.01 0.5 Atom2Vec 多项式核 2 1.0 0.01 2.0 Atom_DL 多项式核 3 1.0 0.15 2.5 表 4 测试集材料预测值和计算值对比
Table 4. Comparison of material predictive and experimental values in the test.
材料
化合物带隙
计算值随机森林 核岭回归 支持向量机 Magpie Atom2Vec Atom_DL Magpie Atom2Vec Atom
_DLMagpie Atom2Vec Atom
_DLClSbTe 1.255 1.198 1.108 1.236 1.176 1.280 1.157 1.253 1.336 1.296 ISSb 1.219 0.885 0.988 1.061 0.849 1.111 1.114 1.068 0.765 1.219 ZrBrI 0.774 0.706 0.665 0.702 0.484 0.700 0.952 0.566 0.856 0.975 ClSbSe 1.172 1.321 1.283 1.343 1.446 1.461 1.282 1.294 1.443 1.397 ZrSSe 0.829 0.569 0.595 0.680 0.596 0.603 0.885 0.578 0.641 0.861 MoSSe 1.453 0.947 0.783 0.932 1.089 0.997 1.394 1.167 1.357 1.220 CrSeTe 0.572 0.258 0.247 0.205 0.382 0.240 0.338 0.349 0.409 0.395 TiClI 0.745 0.601 0.524 0.602 0.408 0.749 0.717 0.554 0.792 0.636 VClI 1.100 0.769 0.726 0.623 0.501 0.714 1.307 0.721 0.990 0.750 VBrCl 1.289 1.081 1.005 1.095 0.633 0.918 1.052 0.915 1.383 1.159 ZrBrCl 0.912 0.971 0.896 0.920 0.764 0.955 1.048 0.920 1.217 1.074 BiIS 0.401 0.698 0.692 0.700 0.541 0.723 0.509 0.659 0.869 0.838 WSTe 1.141 0.646 0.635 0.634 0.695 0.531 1.006 0.940 0.681 0.875 BiClSe 1.235 0.952 0.998 1.127 1.022 0.993 1.204 0.985 0.985 1.085 TiBrCl 0.830 1.106 0.860 0.776 0.536 0.918 0.863 0.751 1.125 0.887 ZrClI 0.877 0.633 0.638 0.633 0.610 0.794 0.982 0.700 0.994 0.772 AsClSe 1.717 1.494 1.549 1.512 1.559 1.433 1.490 1.475 1.579 1.498 AsBrS 1.417 1.447 1.417 1.468 1.342 1.184 1.211 1.465 1.429 1.444 ZrSTe 0.208 0.237 0.218 0.245 0.238 0.145 0.198 0.249 0.076 0.237 ISSb 0.794 0.741 0.817 0.919 0.851 1.113 0.971 1.070 0.944 1.297 BrSbTe 1.319 0.878 1.029 0.983 1.110 1.015 1.256 1.144 1.208 1.041 BiBrS 1.188 0.929 1.067 1.114 1.016 0.858 1.041 0.949 1.184 1.079 ZrSSe 0.613 0.581 0.706 0.766 0.595 0.603 0.538 0.577 0.448 0.651 BiITe 0.346 0.530 0.508 0.481 0.464 0.518 0.033 0.420 0.376 0.173 ClSbTe 1.255 1.198 1.108 1.236 1.176 1.280 1.157 1.253 1.336 1.296 -
[1] He K M, Zhang X Y, Ren S Q, Sun J 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Las Vegas, NV, USA, June 27–30, 2016 p770
[2] Ren S Q, He K M, Girshick R, Sun J 2017 IEEE Trans. Pattern Anal. Mach. Intell. 39 1137
 Google Scholar
Google Scholar
[3] Devlin J, Chang M W, Lee K, Toutanova K 2019 Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies Minneapolis, USA, June 3–5, 2019 p4171
[4] 郭佳龙, 王宗国, 王彦棡, 赵旭山, 宿彦京, 刘志威 2021 数据与计算发展前沿 3 120
 Google Scholar
Google Scholar
Guo J L, Wang Z G, Wang Y G, Zhao X S, Su Y J, Liu Z W 2021 Frontiers of Data and Computing 3 120
 Google Scholar
Google Scholar
[5] 牛程程, 李少波, 胡建军, 但雅波, 曹卓, 李想 2020 材料导报 34 23100
 Google Scholar
Google Scholar
Niu C C, Li S B, Hu J J, Dan Y B, Cao Z, Li X 2020 Mater. Rep. 34 23100
 Google Scholar
Google Scholar
[6] Hu T T, Song H, Jiang T, Li S B 2020 Symmetry 12 1889
 Google Scholar
Google Scholar
[7] Chen C, Ye W K, Zuo Y X, Zheng C, Ong S P 2019 Chem. Mater. 31 3564
 Google Scholar
Google Scholar
[8] Li S B, Dan Y B, Li X, Hu T T, Dong R Z, Cao Z, Hu J J 2020 Symmetry 12 262
 Google Scholar
Google Scholar
[9] Zhang L F, Han J Q, Wang H, Car R, E W N 2018 Phys. Rev. Lett. 120 143001
 Google Scholar
Google Scholar
[10] de Jong M, Chen W, Notestine R, Persson K, Ceder G, Jain A, Asta M, Gamst A 2016 Sci. Rep. 6 34256
 Google Scholar
Google Scholar
[11] Zhou Q, Tang P Z, Liu S X, Pan J B, Yan Q M, Zhang S C 2018 Proc. Nat1. Acad. Sci. U. S. A. 115 6411
 Google Scholar
Google Scholar
[12] Calfa B A, Kitchin J R 2016 AIChE J. 62 2605
 Google Scholar
Google Scholar
[13] Ward L, Agrawal A, Choudhary A, Wolverton C 2016 NPJ Comput. Mater. 2 16028
 Google Scholar
Google Scholar
[14] Zhuo Y, Mansouri Tehrani A, Brgoch J 2018 J. Phys. Chem. Lett. 9 1668
 Google Scholar
Google Scholar
[15] Hu M X, Yuan J M, Sun T, Huang M, Liang Q Y 2021 Comput. Mater. Sci. 200 110841
 Google Scholar
Google Scholar
[16] Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A N, Kaiser Ł, Polosukhin I 2017 31st Conference on Neural Information Processing Systems (NIPS 2017) Long Beach, CA, USA, December 4–9, 2017 p6000
[17] Saal J E, Kirklin S, Aykol M, Meredig B, Wolverton C 2013 JOM 65 1501
 Google Scholar
Google Scholar
[18] Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z M, Gimelshein N, Antiga L, Desmaison A, Köpf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J J, Chintala S 2019 Proceedings of the 33rd International Conference on Neural Information Processing Systems Vancouver, Canada, December 8–14, 2019 p8026
[19] Wang G, Chernikov A, Glazov M M, Heinz T F, Marie X, Amand T, Urbaszek B 2018 Rev. Mod. Phys. 90 021001
 Google Scholar
Google Scholar
[20] Riis-Jensen A C, Deilmann T, Olsen T, Thygesen K S 2019 ACS Nano 13 13354
 Google Scholar
Google Scholar
[21] Gjerding M N, Taghizadeh A, Rasmussen A, Ali S, Bertoldo F, Deilmann T, Knøsgaard N R, Kruse M, Larsen A H, Manti S, Pedersen T G, Petralanda U, Skovhus T, Svendsen M K, Mortensen J J, Olsen T, Thygesen K S 2021 2D Mater. 8 044002
 Google Scholar
Google Scholar
[22] Haastrup S, Strange M, Pandey M, Deilmann T, Schmidt P S, Hinsche N F, Gjerding M N, Torelli D, Larsen P M, Riis-Jensen A C, Gath J, Jacobsen K W, Jørgen Mortensen J, Olsen T, Thygesen K S 2018 2D Mater. 5 042002
 Google Scholar
Google Scholar
[23] Schütt K T, Glawe H, Brockherde F, Sanna A, Müller K R, Gross E K U 2014 Phys. Rev. B 89 205118
 Google Scholar
Google Scholar
[24] Wu Y R, Li H P, Gan X S 2013 Adv. Mater. Res. 848 122
 Google Scholar
Google Scholar
[25] Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É 2011 J. Mach. Learn. Res. 12 2825
 Google Scholar
Google Scholar
计量
- 文章访问数: 8716
- PDF下载量: 122
- 被引次数: 0














 下载:
下载: