-
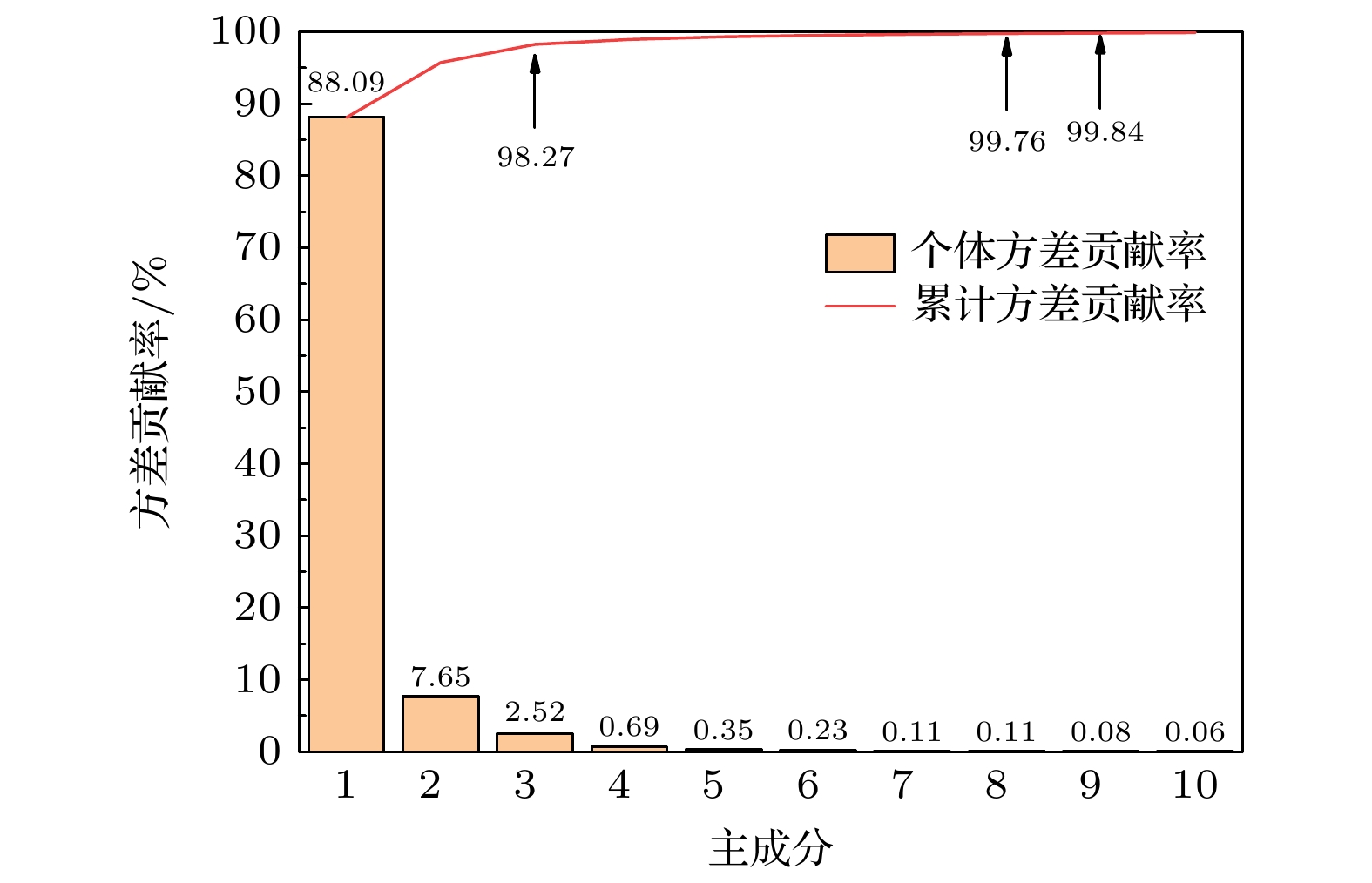
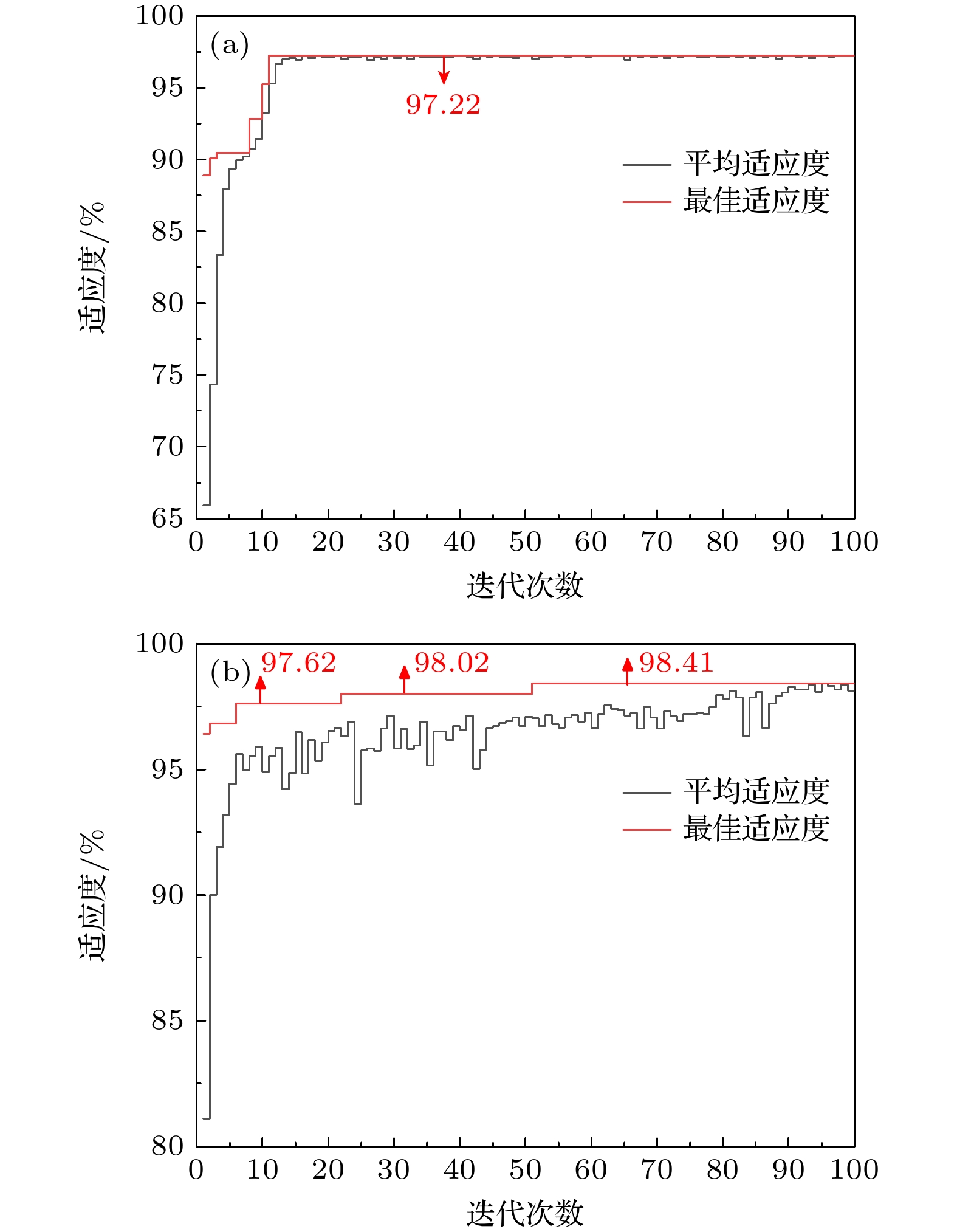
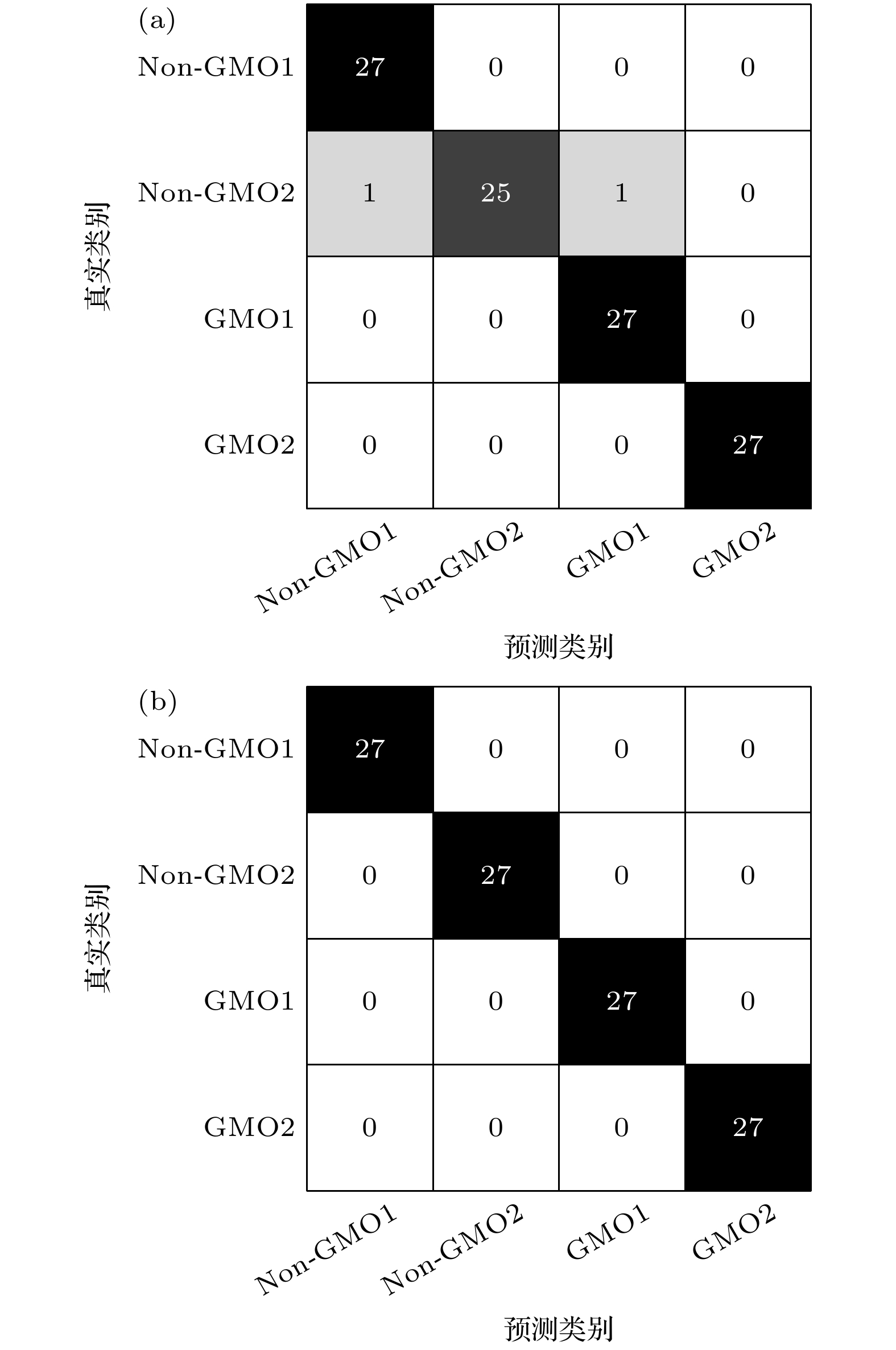
为实现对转基因和非转基因菜籽油的快速准确鉴别, 结合太赫兹时域光谱技术, 提出了一种基于改进蜉蝣优化算法的支持向量机模型. 以两种转基因和两种非转基因菜籽油为研究对象, 应用太赫兹时域光谱技术获取其光谱信息, 发现相比于非转基因菜籽油, 转基因菜籽油在太赫兹波段具有更强的吸收特性, 同时它们的吸收光谱极为相似, 难以通过观察法进行准确区分. 为此, 提出一种基于改进蜉蝣优化算法的支持向量机模型, 通过采用蜉蝣优化算法对支持向量机参数进行寻优, 并引入自适应惯性权重和Lévy飞行两种策略改进蜉蝣优化算法在寻优过程容易陷入局部最优解的问题, 增强蜉蝣优化算法的全局搜索能力和稳健性. 实验结果表明: 改进后的蜉蝣优化算法能够更有效地寻找到支持向量机的最优参数组合, 提升鉴别模型的整体性能, 该模型对4种菜籽油的识别精度为100%. 因此, 本研究为转基因菜籽油的类型鉴别提供了一种快速有效的新方法, 也为其他转基因物质的鉴别提供了有价值的参考.To achieve rapid and accurate identification of genetically modified (GM) and non-GM rapeseed oils, a support vector machine (SVM) model based on an improved mayfly optimization algorithm and coupled with the terahertz time-domain spectroscopy, is proposed. Two types of GM rapeseed oils and two types of non-GM rapeseed oils are selected as research subjects. Their spectral information is acquired by using the terahertz time-domain spectroscopy. The observations show that GM rapeseed oils exhibit stronger terahertz absorption characteristics than non-GM rapeseed oils. However, their absorption spectra are highly similar, making direct differentiation difficult through visual inspection alone. Therefore, SVM is used for spectral recognition. Considering that the classification performance of SVM is significantly affected by its parameters, the mayfly optimization algorithm is combined to optimize these parameters. Furthermore, adaptive inertia weight and Lévy flight strategies are introduced to enhance the global search capability and robustness of the mayfly optimization algorithm, thus addressing the issue of easily becoming trapped in local optima in the optimization process. Moreover, principal component analysis is used to reduce the dimensionality of the absorbance data in a 0.3–1.8 THz range, aiming to extract critical features, thereby enhancing modeling efficiency and reducing redundancy in spectral data. Experimental results demonstrate that the improved mayfly optimization algorithm effectively identifies the optimal parameter combination for SVM, thereby enhancing the overall performance of the identification model. The proposed SVM model, in which the improved mayfly optimization algorithm is used, can achieve a recognition accuracy of 100% for the four types of rapeseed oils, surpassing the 98.15% accuracy achieved by the SVM model with the original mayfly optimization algorithm. Thus, this study presents a rapid and effective new approach for identifying GM rapeseed oils and offers a valuable reference for identifying other genetically modified substances.
-
Keywords:
- transgenic rapeseed oil /
- terahertz spectroscopy /
- classification discrimination /
- mayfly optimization algorithm
[1] 国际农业生物技术应用服务组织 2021 中国生物工程杂志 41 114
ISAAA 2021 China Biotechnol. 41 114
[2] Kumar K, Gambhir G, Dass A, Tripathi A K, Singh A, Jha A K, Yadava P, Choudhary M, Rakshit S 2020 Planta 251 91
 Google Scholar
Google Scholar
[3] Demeke T, Dobnik D 2018 Anal. Bioanal. Chem. 410 4039
 Google Scholar
Google Scholar
[4] Gampala S S, Wulfkuhle B, Richey K A 2019 Transgenic Plants 1864 411
 Google Scholar
Google Scholar
[5] 彭晓昱, 周欢 2021 70 240701
 Google Scholar
Google Scholar
Peng X Y, Zhou H 2021 Acta Phys. Sin. 70 240701
 Google Scholar
Google Scholar
[6] Mittleman D M 2017 J. Appl. Phys. 122 230901
 Google Scholar
Google Scholar
[7] Sun L, Zhao L, Peng R Y 2021 Mil. Med. Res. 8 28
 Google Scholar
Google Scholar
[8] 胡颖, 王晓红, 郭澜涛, 张存林, 刘海波, 张希成 2005 54 4124
 Google Scholar
Google Scholar
Hu Y, Wang X H, Guo L T, Zhang C L, Liu H B, Zhang X C 2005 Acta Phys. Sin. 54 4124
 Google Scholar
Google Scholar
[9] 陈涛 2016 量子电子学报 33 392
Chen T 2016 Chin. J. Quantum Electron. 33 392
[10] 张文涛, 李跃文, 占平平, 熊显名 2017 红外与激光工程 46 1125004
 Google Scholar
Google Scholar
Zhang W T, Li Y W, Zhan P P, Xiong X M 2017 Infrared Laser Eng. 46 1125004
 Google Scholar
Google Scholar
[11] Liu J J 2017 Microw. Opt. Technol. Lett. 59 654
 Google Scholar
Google Scholar
[12] Liu J J, Fan L L, Liu Y M, Mao L L, Kan J Q 2019 Spectrochim. Acta A Mol. Biomol. Spectrosc. 206 165
 Google Scholar
Google Scholar
[13] Gu Q H, Chang Y X, Li X H, Chang Z Z, Feng Z D 2021 Expert Syst. Appl. 165 113713
 Google Scholar
Google Scholar
[14] Guo L, Xu C, Yu T H, Tuerxun W 2022 IEEE Access 10 36335
 Google Scholar
Google Scholar
[15] Cortes C, Vapnik V 1995 Mach. Learn. 20 273
 Google Scholar
Google Scholar
[16] Tuerxun W, Xu C, Guo H Y, Jin Z J, Zhou H J 2021 IEEE Access 9 69307
 Google Scholar
Google Scholar
[17] Zervoudakis K, Tsafarakis S 2020 Comput. Ind. Eng. 145 106559
 Google Scholar
Google Scholar
[18] Ding Y H, You W B 2020 IEEE Access 8 207089
 Google Scholar
Google Scholar
[19] Nickabadi A, Ebadzadeh M M, Safabakhsh R 2011 Appl. Soft Comput. 11 3658
 Google Scholar
Google Scholar
[20] Syama S, Ramprabhakar J, Anand R, Guerrero J M 2023 Results Eng. 19 101274
 Google Scholar
Google Scholar
[21] Liu N, Luo F, Ding W C 2019 2019 IEEE Symposium Series on Computational Intelligence (SSCI) Xiamen, China, December 6–9, 2019 p3104
[22] Pan P Y, Xing Y H, Zhang D W, Wang J, Liu C L, Wu D, Wang X Y 2023 J. Food Sci. 88 3189
 Google Scholar
Google Scholar
[23] Elahi N, Duncan R W, Stasolla C 2016 Plant Physiol. Biochem. 100 52
 Google Scholar
Google Scholar
-
表 1 实验样品信息
Table 1. The information of experimental sample.
标识符 品牌 类型 样本数 训练集 测试集 Non-GMO1 道道全 非转基因 63 27 Non-GMO2 鲁花 非转基因 63 27 GMO1 金龙鱼 转基因 63 27 GMO2 鄉佬坎 转基因 63 27 表 2 两种算法的SVM参数寻优结果
Table 2. Results of SVM parameter optimization under two algorithms.
优化算法 最佳适应度/% 参数 c g MOA 97.22 12.42 0.79 ALMOA 98.41 84.62 0.12 表 3 MOA-SVM模型与ALMOA-SVM模型的性能评价
Table 3. Performance evaluation of the MOA-SVM model and ALMOA-SVM model.
模型 样品 查全率/% 查准率/% 精度/% MOA-SVM Non-GMO1 100 96.43 98.15 Non-GMO2 92.59 100 GMO1 100 96.43 GMO2 100 100 ALMOA-SVM Non-GMO1 100 100 100 Non-GMO2 100 100 GMO1 100 100 GMO2 100 100 -
[1] 国际农业生物技术应用服务组织 2021 中国生物工程杂志 41 114
ISAAA 2021 China Biotechnol. 41 114
[2] Kumar K, Gambhir G, Dass A, Tripathi A K, Singh A, Jha A K, Yadava P, Choudhary M, Rakshit S 2020 Planta 251 91
 Google Scholar
Google Scholar
[3] Demeke T, Dobnik D 2018 Anal. Bioanal. Chem. 410 4039
 Google Scholar
Google Scholar
[4] Gampala S S, Wulfkuhle B, Richey K A 2019 Transgenic Plants 1864 411
 Google Scholar
Google Scholar
[5] 彭晓昱, 周欢 2021 70 240701
 Google Scholar
Google Scholar
Peng X Y, Zhou H 2021 Acta Phys. Sin. 70 240701
 Google Scholar
Google Scholar
[6] Mittleman D M 2017 J. Appl. Phys. 122 230901
 Google Scholar
Google Scholar
[7] Sun L, Zhao L, Peng R Y 2021 Mil. Med. Res. 8 28
 Google Scholar
Google Scholar
[8] 胡颖, 王晓红, 郭澜涛, 张存林, 刘海波, 张希成 2005 54 4124
 Google Scholar
Google Scholar
Hu Y, Wang X H, Guo L T, Zhang C L, Liu H B, Zhang X C 2005 Acta Phys. Sin. 54 4124
 Google Scholar
Google Scholar
[9] 陈涛 2016 量子电子学报 33 392
Chen T 2016 Chin. J. Quantum Electron. 33 392
[10] 张文涛, 李跃文, 占平平, 熊显名 2017 红外与激光工程 46 1125004
 Google Scholar
Google Scholar
Zhang W T, Li Y W, Zhan P P, Xiong X M 2017 Infrared Laser Eng. 46 1125004
 Google Scholar
Google Scholar
[11] Liu J J 2017 Microw. Opt. Technol. Lett. 59 654
 Google Scholar
Google Scholar
[12] Liu J J, Fan L L, Liu Y M, Mao L L, Kan J Q 2019 Spectrochim. Acta A Mol. Biomol. Spectrosc. 206 165
 Google Scholar
Google Scholar
[13] Gu Q H, Chang Y X, Li X H, Chang Z Z, Feng Z D 2021 Expert Syst. Appl. 165 113713
 Google Scholar
Google Scholar
[14] Guo L, Xu C, Yu T H, Tuerxun W 2022 IEEE Access 10 36335
 Google Scholar
Google Scholar
[15] Cortes C, Vapnik V 1995 Mach. Learn. 20 273
 Google Scholar
Google Scholar
[16] Tuerxun W, Xu C, Guo H Y, Jin Z J, Zhou H J 2021 IEEE Access 9 69307
 Google Scholar
Google Scholar
[17] Zervoudakis K, Tsafarakis S 2020 Comput. Ind. Eng. 145 106559
 Google Scholar
Google Scholar
[18] Ding Y H, You W B 2020 IEEE Access 8 207089
 Google Scholar
Google Scholar
[19] Nickabadi A, Ebadzadeh M M, Safabakhsh R 2011 Appl. Soft Comput. 11 3658
 Google Scholar
Google Scholar
[20] Syama S, Ramprabhakar J, Anand R, Guerrero J M 2023 Results Eng. 19 101274
 Google Scholar
Google Scholar
[21] Liu N, Luo F, Ding W C 2019 2019 IEEE Symposium Series on Computational Intelligence (SSCI) Xiamen, China, December 6–9, 2019 p3104
[22] Pan P Y, Xing Y H, Zhang D W, Wang J, Liu C L, Wu D, Wang X Y 2023 J. Food Sci. 88 3189
 Google Scholar
Google Scholar
[23] Elahi N, Duncan R W, Stasolla C 2016 Plant Physiol. Biochem. 100 52
 Google Scholar
Google Scholar
计量
- 文章访问数: 5472
- PDF下载量: 65
- 被引次数: 0














 下载:
下载: