-
DNA聚合酶是执行DNA复制和修复的重要蛋白, 由于其只能从5′向3′方向聚合, 所以在聚合双链DNA时会有两种模式: 其一是先打开DNA双链, 让其暴露出3′-5′方向的模板链(先导链), 然后沿着这条链复制出新链以此置换旧链, 这就是链置换的合成. 另一种是沿着已经置换出的5′-3′方向的模板链(滞后链)进行延伸合成. T7噬菌体作为常被研究的模式生物, 其DNA聚合酶gp5在复制过程中既会参与链置换也会参与延伸合成, 已有的研究报道gp5自身独立链置换的能力很弱, 其与T7解旋酶gp4耦合形成复制体后可以发生快速且持续的链置换, 这一现象的分子机制尚待厘清. 本文通过单分子荧光共振能量转移(smFRET)的方法对gp5聚合过程的动力学进行了研究, 发现gp5在没有外力帮助下时会进入链置换-外切的循环, 导致聚合难以延伸, 调控这一循环的关键则是DNA双链退火压力. 进一步的实验表明gp5和gp4形成复制体后, gp4辅助gp5克服了退火压力从而聚合可以延伸.DNA polymerase is essential for DNA replication and repair. As it only performs the 5′-3′ polymerization, there are two kinds of DNA replication. One of them is called strand-displacement synthesis: DNA polymerase opens the double-strand (ds) DNA to attain the 3′-5′strand (leading strand) and copy this template in a continuous way, and the other is extension synthesis: DNA polymerase copies the newly separated 5′-3′ strand (lagging strand) in a discontinuous manner. The replication complex of T7 phage is an optimal model to investigate the mechanism of replication because it is only constituted by 4 terms of protein which are DNA helicase gp4, DNA polymerase gp5 with co-factor thioredoxin (Trx), and single-strand (ss) DNA-binding protein gp2.5. The replication complex of T7 encounters both strand-displacement synthesis and extension synthesis. Previous researches reported that gp5 can have rapid extension synthesis but lacks the ability to attain strand-displacement synthesis. It also reported that gp4 translocates on ssDNA at a rapid speed but unwinds dsDNA at a very low speed. However, gp5 and gp4 together can attain rapid and processive strand-displacement synthesis. Although extensively studied, this mechanism remains unclear. Here in this work, the dynamic of strand-displacement synthesis by gp5 is investigated with single-molecule Förster (fluorescence) resonance energy transfer (smFRET). It is found that gp5, without the help of external tension, can open dsDNA but only attain strand-displacement synthesis about 4 base pairs (bp), because its exonuclease activity excises the nascent nucleotides. Therefore gp5 repeats in the synthesis-excision cycle which results in the less production of strand-displacement synthesis. We conduct another control experiment by nano-tensioner, a high precision smFRET setup which can exert a tension on dsDNA, to change the dsDNA regression pressure on gp5. It is observed that reduced dsDNA regression pressure can increase the length of strand-displacement synthesis and reduce the length of excision which indicates that the dsDNA regression pressure can regulate the strand-displacement synthesis of gp5. The further experiment shows that after gp5 and gp4 are assembled into a replisome, it can have a processive strand-displacement synthesis and barely any excision presented. The speed of replisome is a little higher than gp5 alone but much higher than gp4 alone. Additionally, the length of strand-displacement synthesis by replisome is much longer than gp5 alone. Therefore it is indicated that the gp4 can reduce dsDNA regression pressure to enables gp5 to attain processive strand-displacement synthesis. On the other hand, the gp5 facilitates gp4 to unwind the dsDNA.
-
Keywords:
- T7 DNA polymerase /
- T7 DNA helicase /
- fluorescence resonance energy transfer /
- DNA regression pressure
[1] Benkovic S J, Valentine A M, Salinas F 2001 Annu. Rev. Biochem. 70 181
 Google Scholar
Google Scholar
[2] O'Donnell M 2006 J. Biol. Chem. 281 10653
 Google Scholar
Google Scholar
[3] Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel S S 2009 Nature 462 940
 Google Scholar
Google Scholar
[4] Sun B, Pandey M, Inman J T, Yang Y, Kashlev M, Patel S S, Wang M D 2015 Nat. Commun. 6 10260
 Google Scholar
Google Scholar
[5] Kath J E, Jergic S, Heltzel J M, Jacob D T, Dixon N E, Sutton M D, Walker G C, Loparo J J 2014 Proc. Natl. Acad. Sci. U.S.A. 111 7647
 Google Scholar
Google Scholar
[6] Nandakumar D, Pandey M, Patel S S 2015 Elife 4 e06562
 Google Scholar
Google Scholar
[7] Pandey M, Patel S S 2014 Cell Rep. 6 1129
 Google Scholar
Google Scholar
[8] Syed S, Pandey M, Patel S S, Ha T 2014 Cell Rep. 6 1037
 Google Scholar
Google Scholar
[9] Gao Y, Cui Y, Fox T, Lin S, Wang H, de Val N, Zhou Z H, Yang W 2019 Science 363 eaav7003
 Google Scholar
Google Scholar
[10] Stano N M, Jeong Y J, Donmez I, Tummalapalli P, Levin M K, Patel S S 2005 Nature 435 370
 Google Scholar
Google Scholar
[11] Manosas M, Spiering M M, Ding F, Bensimon D, Allemand J F, Benkovic S J, Croquette V 2012 Nucleic Acids Res. 40 6174
 Google Scholar
Google Scholar
[12] Lin W, Ma J, Nong D, Xu C, Zhang B, Li J, Jia Q, Dou S, Ye F, Xi X, Lu Y, Li M 2017 Phys. Rev. Lett. 119 138102
 Google Scholar
Google Scholar
[13] Ma J B, Jia Q, Xu C H, Li J H, Huang X Y, Ma D F, Li M, Xi X G, Lu Y 2018 J. Phys. Chem. B 122 5790
 Google Scholar
Google Scholar
[14] 黄星榞, 隋明宇, 侯文清, 李明, 陆颖, 徐春华 2020 69 208706
 Google Scholar
Google Scholar
Huang X Y, Sui M Y, Hou W Q, Li M, Lu Y, Xu C H 2020 Acta Phys. Sin. 69 208706
 Google Scholar
Google Scholar
[15] Kim D E, Narayan M, Patel S S 2002 J. Mol. Biol. 321 807
 Google Scholar
Google Scholar
[16] Ma J B, Chen Z, Xu C H, Huang X Y, Jia Q, Zou Z Y, Mi C Y, Ma D F, Lu Y, Zhang H D, Li M 2020 Nucleic Acids Res. 48 3156
 Google Scholar
Google Scholar
[17] Schwartz J J, Quake S R 2009 Proc. Natl. Acad. Sci. U.S.A. 106 20294
 Google Scholar
Google Scholar
[18] Ha T 2001 Methods 25 78
 Google Scholar
Google Scholar
[19] Etson C M, Hamdan S M, Richardson C C, van Oijen A M 2010 Proc. Natl. Acad. Sci. U.S.A. 107 1900
 Google Scholar
Google Scholar
[20] Ibarra B, Chemla Y R, Plyasunov S, Smith S B, Lazaro J M, Salas M, Bustamante C 2009 EMBO J. 28 2794
 Google Scholar
Google Scholar
[21] Hoekstra T P, Depken M, Lin S N, Cabanas-Danes J, Gross P, Dame R T, Peterman E J G, Wuite G J L 2017 Biophys. J. 112 575
 Google Scholar
Google Scholar
[22] Johansson E, Dixon N 2013 Cold Spring Harb. Perspect. Biol. 5 a012799
 Google Scholar
Google Scholar
[23] Derbyshire V, Freemont P S, Sanderson M R, Beese L, Friedman J M, Joyce C M, Steitz T A 1988 Science 240 199
 Google Scholar
Google Scholar
[24] Lam W C, Van der Schans E J, Joyce C M, Millar D P 1998 Biochemistry 37 1513
 Google Scholar
Google Scholar
[25] Graham J E, Marians K J, Kowalczykowski S C 2017 Cell 169 1201
 Google Scholar
Google Scholar
[26] 陈泽, 马建兵, 黄星榞, 贾棋, 徐春华, 张慧东, 陆颖 2018 67 118201
 Google Scholar
Google Scholar
Chen Z, Ma J-B, Huang X Y, Jia Q, Xu C H, Zhang H D, Lu Y 2018 Acta Phys. Sin. 67 118201
 Google Scholar
Google Scholar
-
图 1 exo+ gp5和exo– gp5的PAGE实验 (a)电泳实验的DNA, 在引物链5′端标记有Alexa488; (b) 对Alexa488照明得到的结果: 第1个条带为DNA的原始长度, 第2个条带是exo+ gp5合成1 min后的结果, 第3个条带是exo– gp5合成1 min后的结果, 第4个条带是exo+ gp5合成5 min后的结果, 第5个条带是exo– gp5合成5 min后的结果
Fig. 1. PAGE assays of exo+ gp5 and exo– gp5. (a) Illustration of DNA that used in PAGE assays. Alexa488 is labeled on the 5′ overhand of primer DNA. (b) Results of various condition. The first lane: Original length of the DNA. The second lane: The synthesis-product of exo+ gp5 with 1 minute. The third lane: The synthesis-product of exo– gp5 with 1 minute. The fourth lane: The synthesis-product of exo+ gp5 with 5 minute. The fifth lane: The synthesis-product of exo– gp5 with 5 minute.
图 2 T7 DNA聚合酶gp5不断重复链置换和外切 (a), (b)没有结合辅助因子Trx时聚合酶无法链置换DNA双链; (c), (d)有辅助因子Trx时聚合酶能够部分链置换DNA, 但是会回退; (e), (f)外切活性突变后的gp5不会再外切; (g) exo+ gp5 + Trx实验中合成长度的统计图, 其分布满足单e指数; (h) exo+ gp5 + Trx实验中外切长度的统计图, 其分布满足单e指数
Fig. 2. T7 DNA polymerase gp5 repeats in synthesis-excision cycle: (a), (b) exo+ gp5 cannot have displacement synthesis without co-factor Trx; (c), (d) gp5 with Trx repeats in synthesis-excision cycle; (e), (f) exo– gp5 attain full-length displacement synthesis without excision; (g) histogram of synthesis processivity from assay of exo+ gp5 + Trx, the distribution is well fit by an exponential; (h) histogram of excision processivity from assay of exo+ gp5+ Trx, the distribution is well fit by an exponential.
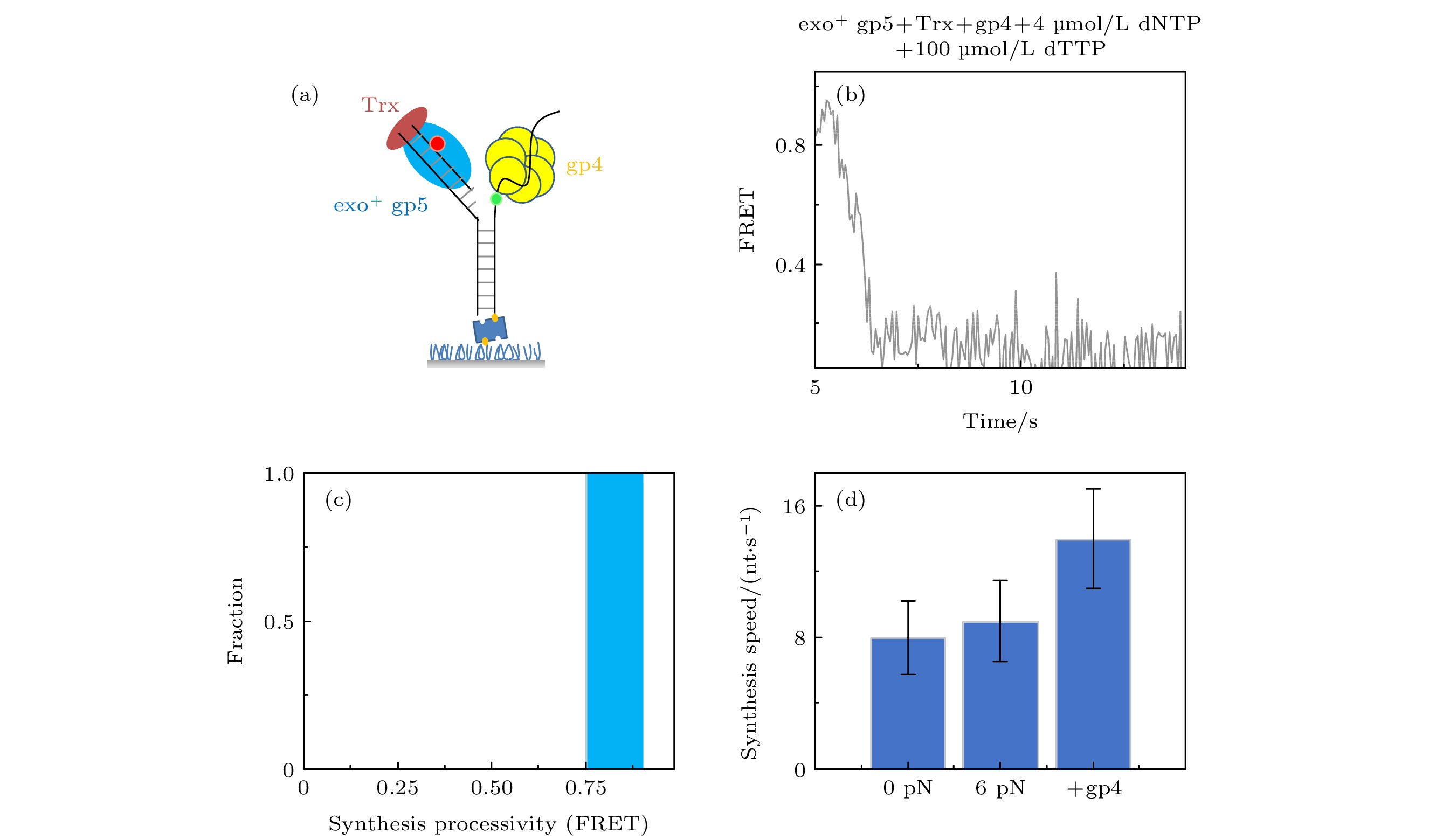
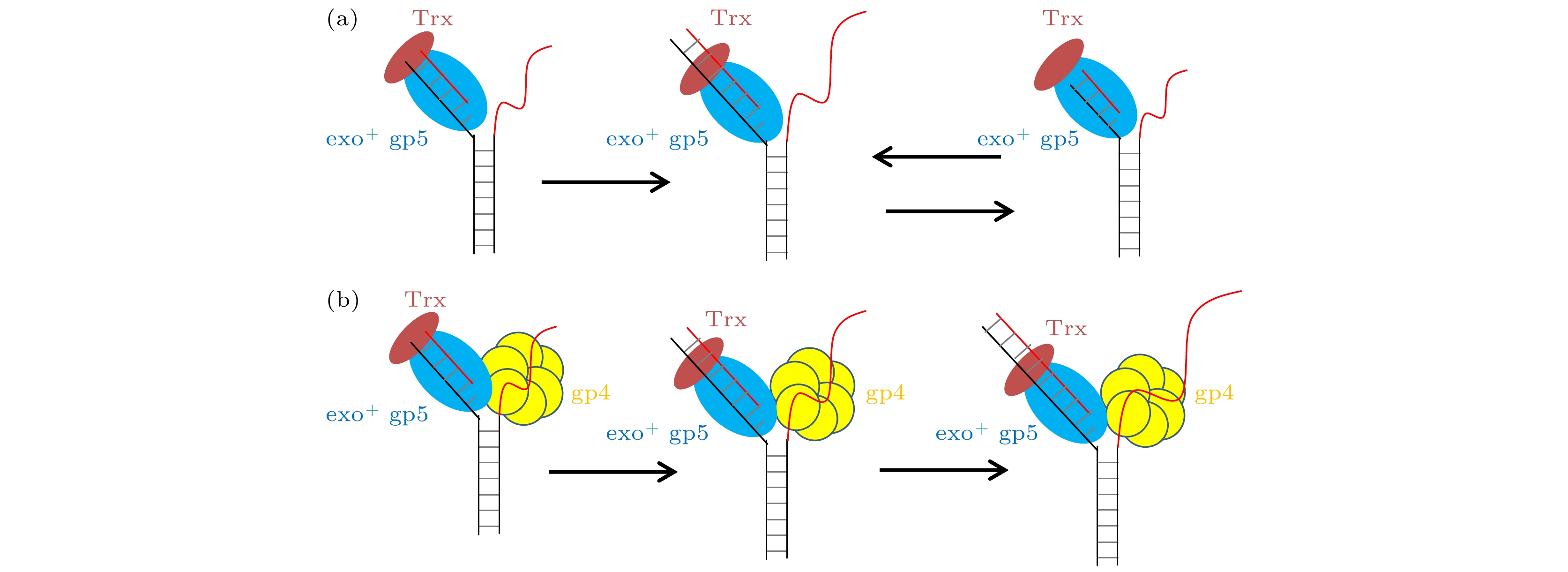
图 4 T7 DNA解旋酶帮助聚合酶克服退火压力 (a) gp4, exo+ gp5和Trx共同链置换的示意图, (b) gp4, exo+ gp5和Trx共同链置换时的典型曲线; (c) exo+ gp5 + Trx + gp4可以完全链置换; (d)不同情况的链置换速度
Fig. 4. gp4 decrease DNA regression pressure which facilitate gp5 to attain processive strand-displacement synthesis: (a) Illustration of displacement by gp4, exo+ gp5 and Trx; (b) typical trace from assay of gp4, exo+ gp5 and Trx; (c) exo+ gp5 + Trx + gp4 attain processive synthesis; (d) synthesis speed in various condition.
图 3 T7 DNA聚合酶gp5外切的原因是退火压力 (a)纳米张力器示意图; (b) 纳米张力器实验的典型曲线; (c) exo+ gp5 + Trx在受力后合成长度的统计图, 其分布满足单e指数; (d) exo+ gp5 + Trx受力后外切长度的统计图, 其分布满足单e指数
Fig. 3. DNA regression pressure induced exonuclease activity: (a) Illustration of nanotensionior; (b) typical trace from assay of nanotensionior; (c) histogram of synthesis processivity from assay of exo+ gp5 + Trx with tension, the distribution is well fit by an exponential; (d) histogram of excision processivity from assay of exo+ gp5 + Trx with tension, the distribution is well fit by an exponential.
-
[1] Benkovic S J, Valentine A M, Salinas F 2001 Annu. Rev. Biochem. 70 181
 Google Scholar
Google Scholar
[2] O'Donnell M 2006 J. Biol. Chem. 281 10653
 Google Scholar
Google Scholar
[3] Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel S S 2009 Nature 462 940
 Google Scholar
Google Scholar
[4] Sun B, Pandey M, Inman J T, Yang Y, Kashlev M, Patel S S, Wang M D 2015 Nat. Commun. 6 10260
 Google Scholar
Google Scholar
[5] Kath J E, Jergic S, Heltzel J M, Jacob D T, Dixon N E, Sutton M D, Walker G C, Loparo J J 2014 Proc. Natl. Acad. Sci. U.S.A. 111 7647
 Google Scholar
Google Scholar
[6] Nandakumar D, Pandey M, Patel S S 2015 Elife 4 e06562
 Google Scholar
Google Scholar
[7] Pandey M, Patel S S 2014 Cell Rep. 6 1129
 Google Scholar
Google Scholar
[8] Syed S, Pandey M, Patel S S, Ha T 2014 Cell Rep. 6 1037
 Google Scholar
Google Scholar
[9] Gao Y, Cui Y, Fox T, Lin S, Wang H, de Val N, Zhou Z H, Yang W 2019 Science 363 eaav7003
 Google Scholar
Google Scholar
[10] Stano N M, Jeong Y J, Donmez I, Tummalapalli P, Levin M K, Patel S S 2005 Nature 435 370
 Google Scholar
Google Scholar
[11] Manosas M, Spiering M M, Ding F, Bensimon D, Allemand J F, Benkovic S J, Croquette V 2012 Nucleic Acids Res. 40 6174
 Google Scholar
Google Scholar
[12] Lin W, Ma J, Nong D, Xu C, Zhang B, Li J, Jia Q, Dou S, Ye F, Xi X, Lu Y, Li M 2017 Phys. Rev. Lett. 119 138102
 Google Scholar
Google Scholar
[13] Ma J B, Jia Q, Xu C H, Li J H, Huang X Y, Ma D F, Li M, Xi X G, Lu Y 2018 J. Phys. Chem. B 122 5790
 Google Scholar
Google Scholar
[14] 黄星榞, 隋明宇, 侯文清, 李明, 陆颖, 徐春华 2020 69 208706
 Google Scholar
Google Scholar
Huang X Y, Sui M Y, Hou W Q, Li M, Lu Y, Xu C H 2020 Acta Phys. Sin. 69 208706
 Google Scholar
Google Scholar
[15] Kim D E, Narayan M, Patel S S 2002 J. Mol. Biol. 321 807
 Google Scholar
Google Scholar
[16] Ma J B, Chen Z, Xu C H, Huang X Y, Jia Q, Zou Z Y, Mi C Y, Ma D F, Lu Y, Zhang H D, Li M 2020 Nucleic Acids Res. 48 3156
 Google Scholar
Google Scholar
[17] Schwartz J J, Quake S R 2009 Proc. Natl. Acad. Sci. U.S.A. 106 20294
 Google Scholar
Google Scholar
[18] Ha T 2001 Methods 25 78
 Google Scholar
Google Scholar
[19] Etson C M, Hamdan S M, Richardson C C, van Oijen A M 2010 Proc. Natl. Acad. Sci. U.S.A. 107 1900
 Google Scholar
Google Scholar
[20] Ibarra B, Chemla Y R, Plyasunov S, Smith S B, Lazaro J M, Salas M, Bustamante C 2009 EMBO J. 28 2794
 Google Scholar
Google Scholar
[21] Hoekstra T P, Depken M, Lin S N, Cabanas-Danes J, Gross P, Dame R T, Peterman E J G, Wuite G J L 2017 Biophys. J. 112 575
 Google Scholar
Google Scholar
[22] Johansson E, Dixon N 2013 Cold Spring Harb. Perspect. Biol. 5 a012799
 Google Scholar
Google Scholar
[23] Derbyshire V, Freemont P S, Sanderson M R, Beese L, Friedman J M, Joyce C M, Steitz T A 1988 Science 240 199
 Google Scholar
Google Scholar
[24] Lam W C, Van der Schans E J, Joyce C M, Millar D P 1998 Biochemistry 37 1513
 Google Scholar
Google Scholar
[25] Graham J E, Marians K J, Kowalczykowski S C 2017 Cell 169 1201
 Google Scholar
Google Scholar
[26] 陈泽, 马建兵, 黄星榞, 贾棋, 徐春华, 张慧东, 陆颖 2018 67 118201
 Google Scholar
Google Scholar
Chen Z, Ma J-B, Huang X Y, Jia Q, Xu C H, Zhang H D, Lu Y 2018 Acta Phys. Sin. 67 118201
 Google Scholar
Google Scholar
计量
- 文章访问数: 8832
- PDF下载量: 96
- 被引次数: 0














 下载:
下载: