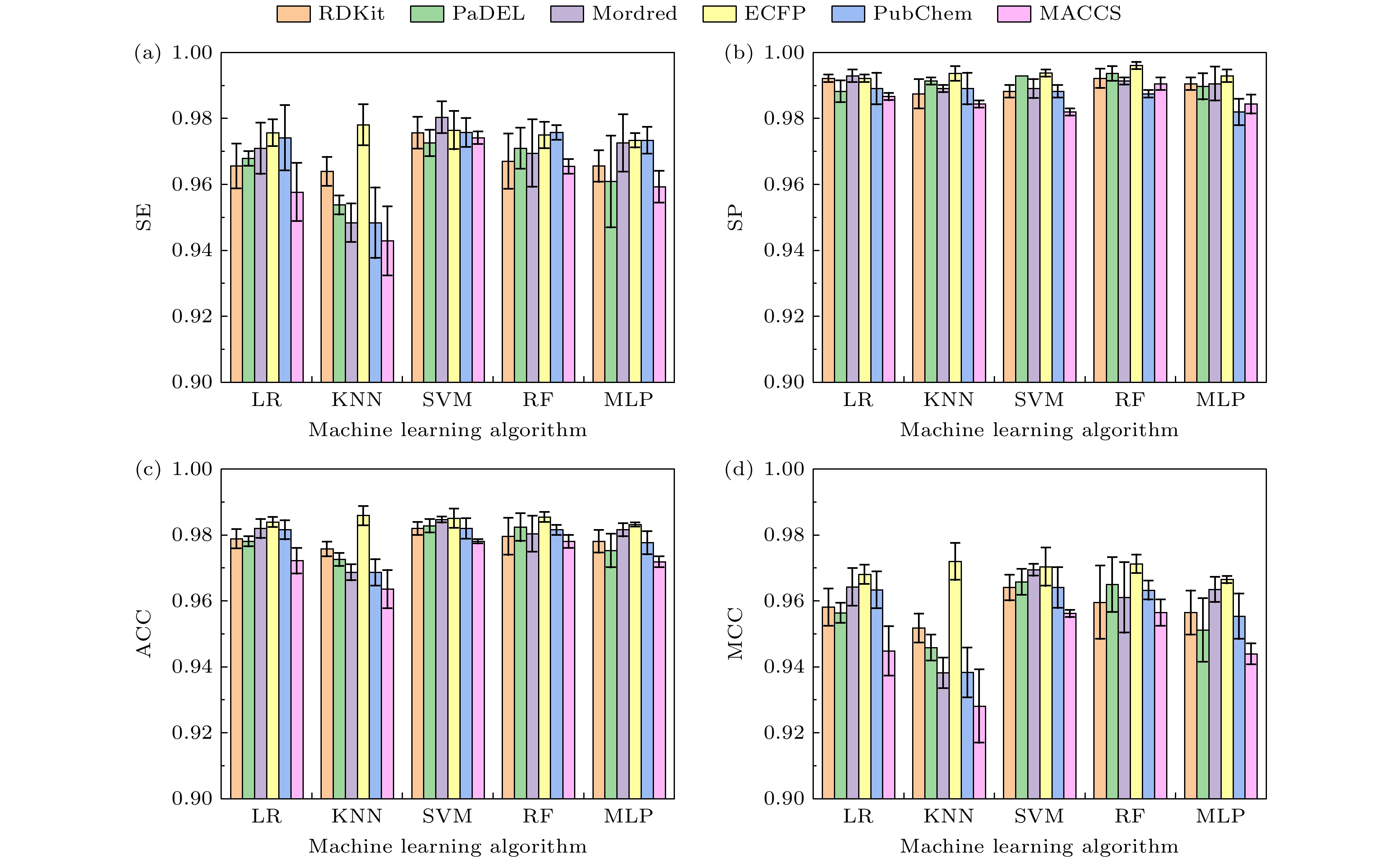
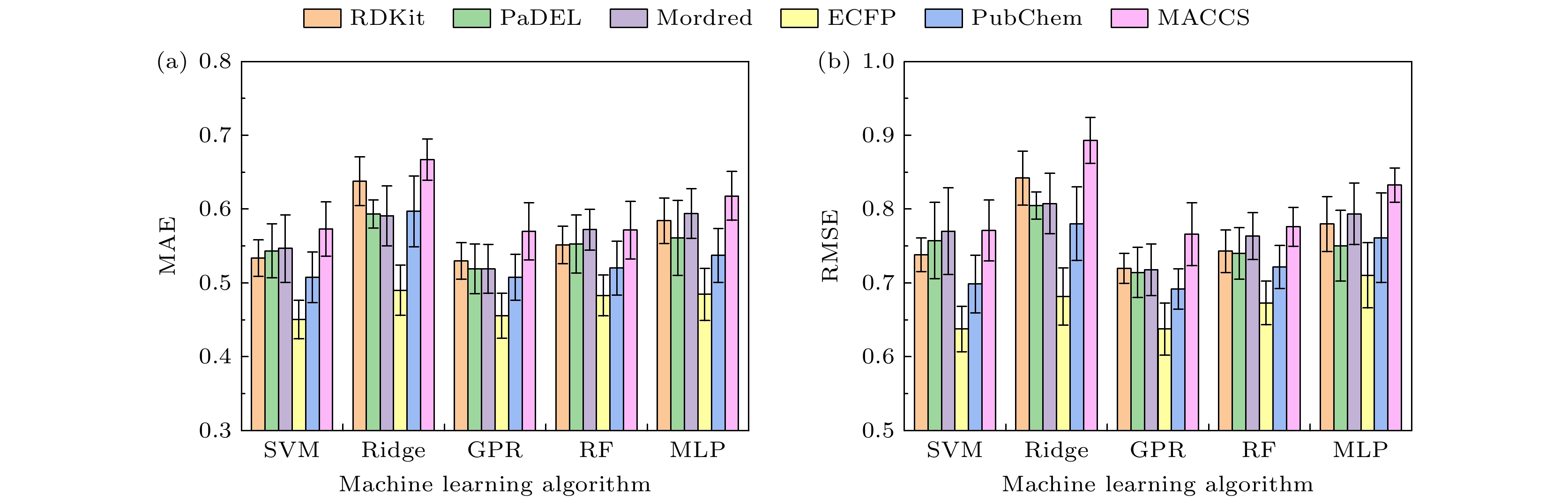
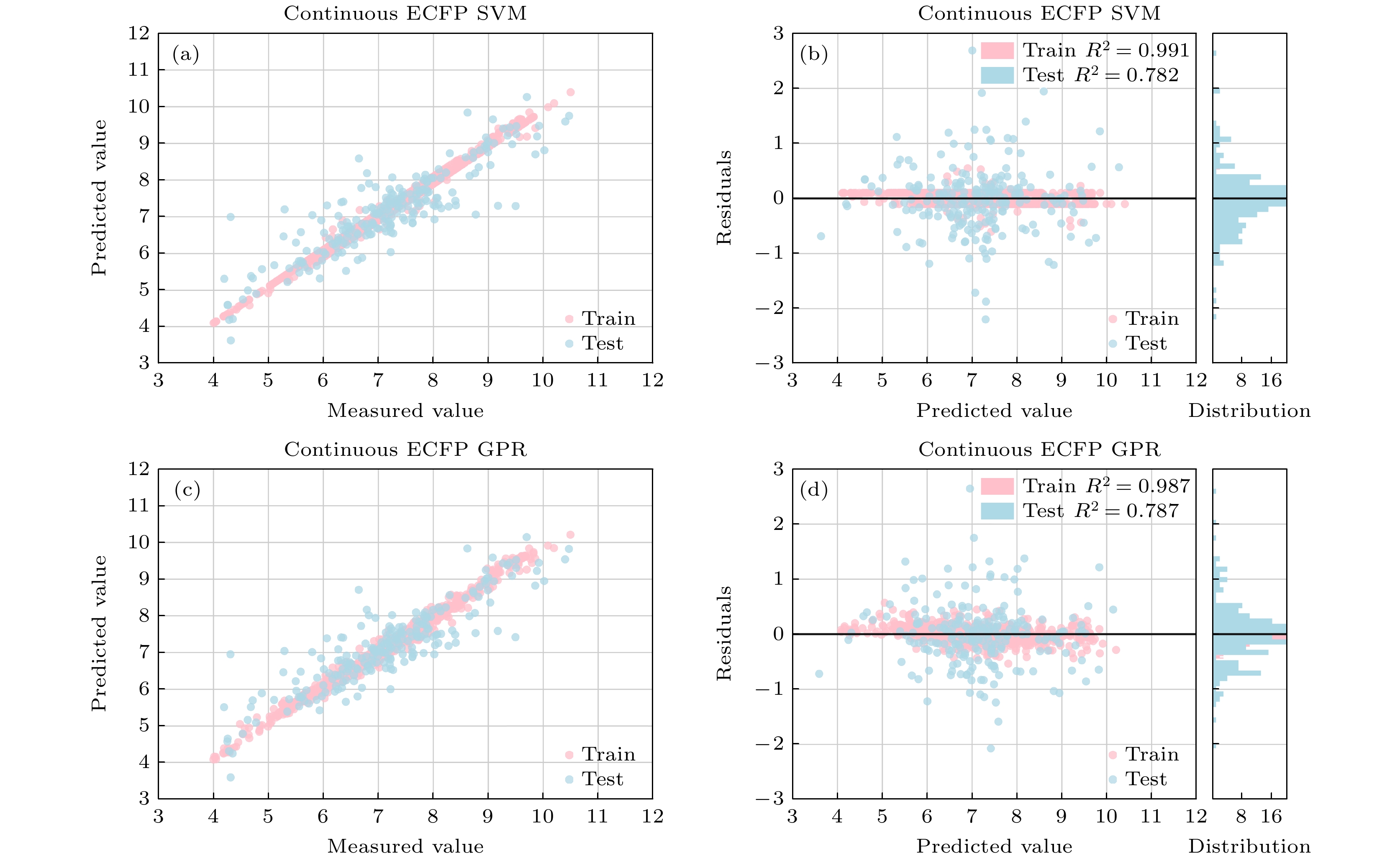
-
针对PD-1/PD-L1免疫检查点的单克隆抗体抑制剂逐渐进入市场并在多种类型的肿瘤治疗中取得一定的积极效果. 然而, 随着应用范围的不断扩展, 抗体药物的局限性以及过多同质化研究等问题逐渐显现出来, 小分子化合物抑制剂成为了研究者们关注的新焦点. 本文旨在利用基于配体和基于结构的结合活性预测方法实现针对PD-L1靶点的小分子化合物虚拟筛选, 从而帮助加速小分子药物的开发. 通过从相关研究文献及专利收集PD-L1小分子抑制活性数据集, 根据不同分子表征方法和算法构建机器学习活性判定分类模型和活性强度预测回归模型, 两类模型从大型类药小分子库(ZINC15)中筛选获得68种高PD-L1抑制活性候选化合物. 其中10种化合物不仅具备良好的药物相似性和药代动力学, 还在分子对接中与已报道的热点化合物表现出同等水平的结合强度和相似的作用机制, 这一现象在后续分子动力学模拟和结合自由能估计中得到进一步验证. 本文提出了一个融合基于配体方法和基于结构方法的计算机辅助药物研发工作流程, 其在大型化合物数据库中有效筛选出有潜力的PD-L1小分子抑制剂, 有望助力加速肿瘤免疫治疗的应用.
-
关键词:
- PD-1/PD-L1 /
- 虚拟筛选 /
- 机器学习 /
- 分子动力学模拟
Monoclonal antibody inhibitors targeting PD-1/PD-L1 immune checkpoints are gradually entering the market and have achieved certain positive effects in the treatments of various types of tumors. However, with the expansion of application, the limitations of antibody drugs and problems such as excessive homogenization of research gradually appear, making small-molecule inhibitors the new focus of researchers. This study aims to use ligand-based and structure-based binding activity prediction methods to achieve virtual screening of small-molecule inhibitors targeting PD-L1, thereby helping to accelerate the development of small molecule drugs. A dataset of PD-L1 small-molecule inhibitory activity from relevant research literature and patents is collected and activity judgment classification models with intensity prediction regression models are constructed based on different molecular featurization methods and machine learning algorithms. The two types of models filter 68 candidate compounds with high PD-L1 inhibitory activity from a large drug-like small molecule screening pool (ZINC15). Ten of these compounds not only have good drug similarities and pharmacokinetics, but also exhibit comparable binding affinities and similar mechanisms of action with previous reported hotspot compounds in molecular docking. This phenomenon is further verified in subsequent molecular dynamics simulation and the estimation of binding free energy. In this study, a virtual screening workflow integrating ligand-based method and structure-based method is developed, and potential PD-L1 small-molecule inhibitors are effectively screened from large compound databases, which is expected to help accelerate the application and expansion of tumor immunotherapy.-
Keywords:
- PD-1/PD-L1 /
- virtual screening /
- machine learning /
- molecular dynamics simulation
[1] Waldman A D, Fritz J M, Lenardo M J 2020 Nat. Rev. Immunol. 20 651
 Google Scholar
Google Scholar
[2] Wherry E J, Kurachi M 2015 Nat. Rev. Immunol. 15 486
 Google Scholar
Google Scholar
[3] Zou W, Wolchok J D, Chen L 2016 Sci. Transl. Med. 8 328rv4
 Google Scholar
Google Scholar
[4] Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, Wu X, Ma J, Zhou M, Li X, Li Y, Li G, Xiong W, Guo C, Zeng Z 2019 Mol. Cancer 18 10
 Google Scholar
Google Scholar
[5] Topalian S L, Drake C G, Pardoll D M 2015 Cancer Cell 27 450
 Google Scholar
Google Scholar
[6] Postow M A, Callahan M K, Wolchok J D 2015 J. Clin. Oncol. 33 1974
 Google Scholar
Google Scholar
[7] Tang Q, Chen Y, Li X, Long S, Shi Y, Yu Y, Wu W, Han L, Wang S 2022 Front. Immunol. 13 964442
 Google Scholar
Google Scholar
[8] Ai L, Xu A, Xu J 2020 Adv. Exp. Med. Biol. 1248 33
 Google Scholar
Google Scholar
[9] Dermani F K, Samadi P, Rahmani G, Kohlan A K, Najafi R 2019 J. Cell. Physiol. 234 1313
 Google Scholar
Google Scholar
[10] Parry R V, Chemnitz J M, Frauwirth K A, Lanfranco A R, Braunstein I, Kobayashi S V, Linsley P S, Thompson C B, Riley J L 2005 Mol. Cell. Biol. 25 9543
 Google Scholar
Google Scholar
[11] Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis V A 2012 Sci. Signal. 5 ra46
 Google Scholar
Google Scholar
[12] Hui E, Cheung J, Zhu J, Su X, Taylor M J, Wallweber H A, Sasmal D K, Huang J, Kim J M, Mellman I, Vale R D 2017 Science 355 1428
 Google Scholar
Google Scholar
[13] Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N 2002 Proc. Natl. Acad. Sci. U.S.A. 99 12293
 Google Scholar
Google Scholar
[14] Iwai Y, Terawaki S, Honjo T 2005 Int. Immunol. 17 133
 Google Scholar
Google Scholar
[15] Gong J, Chehrazi-Raffle A, Reddi S, Salgia R 2018 J. Immunother. Cancer 6 8
 Google Scholar
Google Scholar
[16] Akinleye A, Rasool Z 2019 J. Hematol. Oncol. 12 92
 Google Scholar
Google Scholar
[17] Shiravand Y, Khodadadi F, Kashani S M A, Hosseini-Fard S R, Hosseini S, Sadeghirad H, Ladwa R, O'Byrne K, Kulasinghe A 2022 Curr. Oncol. 29 3044
 Google Scholar
Google Scholar
[18] Zhang J, Zhang Y, Qu B, Yang H, Hu S, Dong X 2021 Eur. J. Med. Chem. 218 113356
 Google Scholar
Google Scholar
[19] Johnson D B, Nebhan C A, Moslehi J J, Balko J M 2022 Nat. Rev. Clin. Oncol. 19 254
 Google Scholar
Google Scholar
[20] Mould D R, Meibohm B 2016 BioDrugs 30 275
 Google Scholar
Google Scholar
[21] Wu Q, Jiang L, Li S C, He Q J, Yang B, Cao J 2021 Acta Pharmacol. Sin. 42 1
 Google Scholar
Google Scholar
[22] Zhan M M, Hu X Q, Liu X X, Ruan B F, Xu J, Liao C 2016 Drug Discov. Today 21 1027
 Google Scholar
Google Scholar
[23] Liu C, Seeram N P, Ma H 2021 Cancer Cell Int. 21 239
 Google Scholar
Google Scholar
[24] Chupak L S, Zheng X 2015 WO Patent 2015034820 A1
[25] Zak K M, Grudnik P, Guzik K, Zieba B J, Musielak B, Dömling A, Dubin G, Holak T A 2016 Oncotarget 7 30323
 Google Scholar
Google Scholar
[26] Koblish H K, Wu L, Wang L S, Liu P C C, Wynn R, Rios-Doria J, Spitz S, Liu H, Volgina A, Zolotarjova N, Kapilashrami K, Behshad E, Covington M, Yang Y O, Li J, Diamond S, Soloviev M, O'Hayer K, Rubin S, Kanellopoulou C, Yang G, Rupar M, DiMatteo D, Lin L, Stevens C, Zhang Y, Thekkat P, Geschwindt R, Marando C, Yeleswaram S, Jackson J, Scherle P, Huber R, Yao W, Hollis G 2022 Cancer Discov. 12 1482
 Google Scholar
Google Scholar
[27] Jiang J, Zou X, Liu Y, Liu X, Dong K, Yao X, Feng Z, Chen X, Sheng L, Li Y 2021 Front. Pharmacol. 12 677120
 Google Scholar
Google Scholar
[28] Wang Y, Liu X, Zou X, Wang S, Luo L, Liu Y, Dong K, Yao X, Li Y, Chen X, Sheng L 2021 Pharmaceutics 13 598
 Google Scholar
Google Scholar
[29] Sobral P S, Luz V C C, Almeida J, Videira P A, Pereira F 2023 Int. J. Mol. Sci. 24 5908
 Google Scholar
Google Scholar
[30] Luo L, Zhong A, Wang Q, Zheng T 2021 Mar. Drugs 20 29
 Google Scholar
Google Scholar
[31] Acúrcio R C, Pozzi S, Carreira B, Pojo M, Gómez-Cebrián N, Casimiro S, Fernandes A, Barateiro A, Farricha V, Brito J, Leandro A P, Salvador J A R, Graça L, Puchades-Carrasco L, Costa L, Satchi-Fainaro R, Guedes R C, Florindo H F 2022 J. Immunother. Cancer 10 e004695
 Google Scholar
Google Scholar
[32] Lung J, Hung M S, Lin Y C, Hung C H, Chen C C, Lee K D, Tsai Y H 2020 Molecules 25 5293
 Google Scholar
Google Scholar
[33] Guo Y, Liang J, Liu B, Jin Y 2021 Int. J. Mol. Sci. 22 10924
 Google Scholar
Google Scholar
[34] Sterling T, Irwin J J 2015 J. Chem. Inf. Model. 55 2324
 Google Scholar
Google Scholar
[35] Bemis G W, Murcko M A 1996 J. Med. Chem. 39 2887
 Google Scholar
Google Scholar
[36] Rogers D, Hahn M 2010 J. Chem. Inf. Model. 50 742
 Google Scholar
Google Scholar
[37] Yap C W 2011 J. Comput. Chem. 32 1466
 Google Scholar
Google Scholar
[38] Durant J L, Leland B A, Henry D R, Nourse J G 2002 J. Chem. Inf. Comput. Sci. 42 1273
 Google Scholar
Google Scholar
[39] Moriwaki H, Tian Y S, Kawashita N, Takagi T 2018 J. Cheminform. 10 4
 Google Scholar
Google Scholar
[40] Bickerton G R, Paolini G V, Besnard J, Muresan S, Hopkins A L 2012 Nat. Chem. 4 90
 Google Scholar
Google Scholar
[41] Kosugi T, Ohue M 2021 Int. J. Mol. Sci. 22 10925
 Google Scholar
Google Scholar
[42] Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D 2021 Nucleic Acids Res. 49 W5
 Google Scholar
Google Scholar
[43] Jing T, Zhang Z, Kang Z, Mo J, Yue X, Lin Z, Fu X, Liu C, Ma H, Zhang X, Hu W 2023 J. Med. Chem. 66 6811
 Google Scholar
Google Scholar
[44] Qin M, Meng Y, Yang H, Liu L, Zhang H, Wang S, Liu C, Wu X, Wu D, Tian Y, Hou Y, Zhao Y, Liu Y, Xu C, Wang L 2021 J. Med. Chem. 64 5519
 Google Scholar
Google Scholar
[45] Trott O, Olson A J 2010 J. Comput. Chem. 31 455
 Google Scholar
Google Scholar
[46] Laskowski R A, Swindells M B 2011 J. Chem. Inf. Model. 51 2778
 Google Scholar
Google Scholar
[47] Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell Jr. A D 2010 J. Comput. Chem. 31 671
 Google Scholar
Google Scholar
[48] Yu W, He X, Vanommeslaeghe K, MacKerell Jr. A D 2012 J. Comput. Chem. 33 2451
 Google Scholar
Google Scholar
[49] Vanommeslaeghe K, MacKerell Jr. A D 2012 J. Chem. Inf. Model. 52 3144
 Google Scholar
Google Scholar
[50] Vanommeslaeghe K, Raman E P, MacKerell Jr. A D 2012 J. Chem. Inf. Model. 52 3155
 Google Scholar
Google Scholar
[51] Abraham M J, Murtola T, Schulz R, Páll S, Smith J C, Hess B, Lindahl E 2015 SoftwareX 1 19
 Google Scholar
Google Scholar
[52] Parrinello M, Rahman A 1981 J. Appl. Phys. 52 7182
 Google Scholar
Google Scholar
[53] Nosé S 1984 Mol. Phys. 52 255
 Google Scholar
Google Scholar
[54] Hoover W G 1985 Phys. Rev. A 31 1695
 Google Scholar
Google Scholar
[55] Essmann U, Perera L, Berkowitz M L, Darden T, Lee H, Pedersen L G 1995 J. Chem. Phys. 103 8577
 Google Scholar
Google Scholar
[56] Hess B, Bekker H, Berendsen H J C, Fraaije J G E M 1997 J. Comput. Chem. 18 1463
 Google Scholar
Google Scholar
[57] Valdés-Tresanco M S, Valdés-Tresanco M E, Valiente P A, Moreno E 2021 J. Chem. Theory Comput. 17 6281
 Google Scholar
Google Scholar
[58] Chupak L S, Ding M, Martin S W, Zheng X, Hewawasam P, Connolly T P, Xu N, Yeung K S, Zhu J, Langley D R, Tenney D J, Scola P M 2015 WO Patent 2015160641 A3
[59] Wang T, Cai S, Wang M, Zhang W, Zhang K, Chen D, Li Z, Jiang S 2021 J. Med. Chem. 64 7390
 Google Scholar
Google Scholar
[60] Lu L, Qi Z, Wang T, Zhang X, Zhang K, Wang K, Cheng Y, Xiao Y, Li Z, Jiang S 2022 ACS Med. Chem. Lett. 13 586
 Google Scholar
Google Scholar
[61] Dai X, Wang K, Chen H, Huang X, Feng Z 2021 Bioorg. Chem. 114 105034
 Google Scholar
Google Scholar
[62] Le Biannic R, Magnez R, Klupsch F, Leleu-Chavain N, Thiroux B, Tardy M, El Bouazzati H, Dezitter X, Renault N, Vergoten G, Bailly C, Quesnel B, Thuru X, Millet R 2022 Eur. J. Med. Chem. 236 114343
 Google Scholar
Google Scholar
[63] Russomanno P, Assoni G, Amato J, D'Amore V M, Scaglia R, Brancaccio D, Pedrini M, Polcaro G, La Pietra V, Orlando P, Falzoni M, Cerofolini L, Giuntini S, Fragai M, Pagano B, Donati G, Novellino E, Quintavalle C, Condorelli G, Sabbatino F, Seneci P, Arosio D, Pepe S, Marinelli L 2021 J. Med. Chem. 64 16020
 Google Scholar
Google Scholar
[64] Liu L, Yao Z, Wang S, Xie T, Wu G, Zhang H, Zhang P, Wu Y, Yuan H, Sun H 2021 J. Med. Chem. 64 8391
 Google Scholar
Google Scholar
[65] Cheng B, Ren Y, Cao H, Chen J 2020 Eur. J. Med. Chem. 199 112377
 Google Scholar
Google Scholar
[66] Lipinski C A 2004 Drug Discov. Today Technol. 1 337
 Google Scholar
Google Scholar
[67] Liang J, Wang B, Yang Y, Liu B, Jin Y 2023 Int. J. Mol. Sci. 24 1280
 Google Scholar
Google Scholar
-
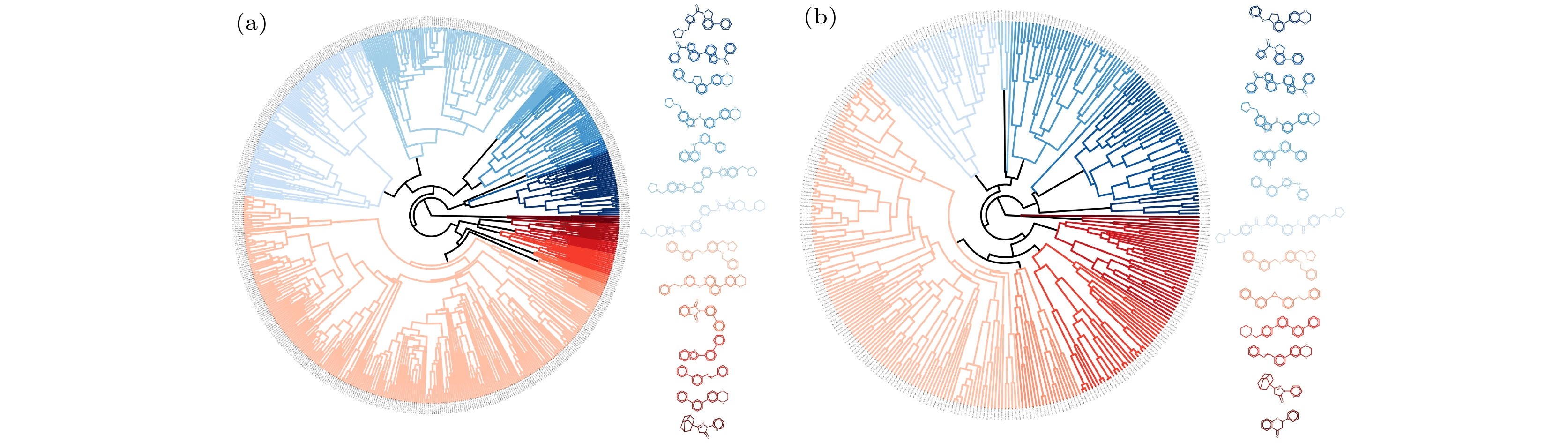
图 1 数据集聚类 (a)分类模型阳性样本骨架的14个簇; (b)回归模型样本骨架的13个簇. 环形树状图的颜色代表不同的分子结构骨架簇, 颜色越接近, 骨架越相似, 一旁的化学结构图是每个簇中最具代表性的骨架
Fig. 1. Dataset clustering: (a) 14 clusters of scaffolds of active compounds in the classification models; (b) 13 clusters of scaffolds of compounds in the regression models. The colors of the circular dendrograms represent different clusters of scaffolds, and the closer the colors are, the more similar the scaffolds are. The chemical structure diagrams on the side are the most representative scaffolds of each cluster.
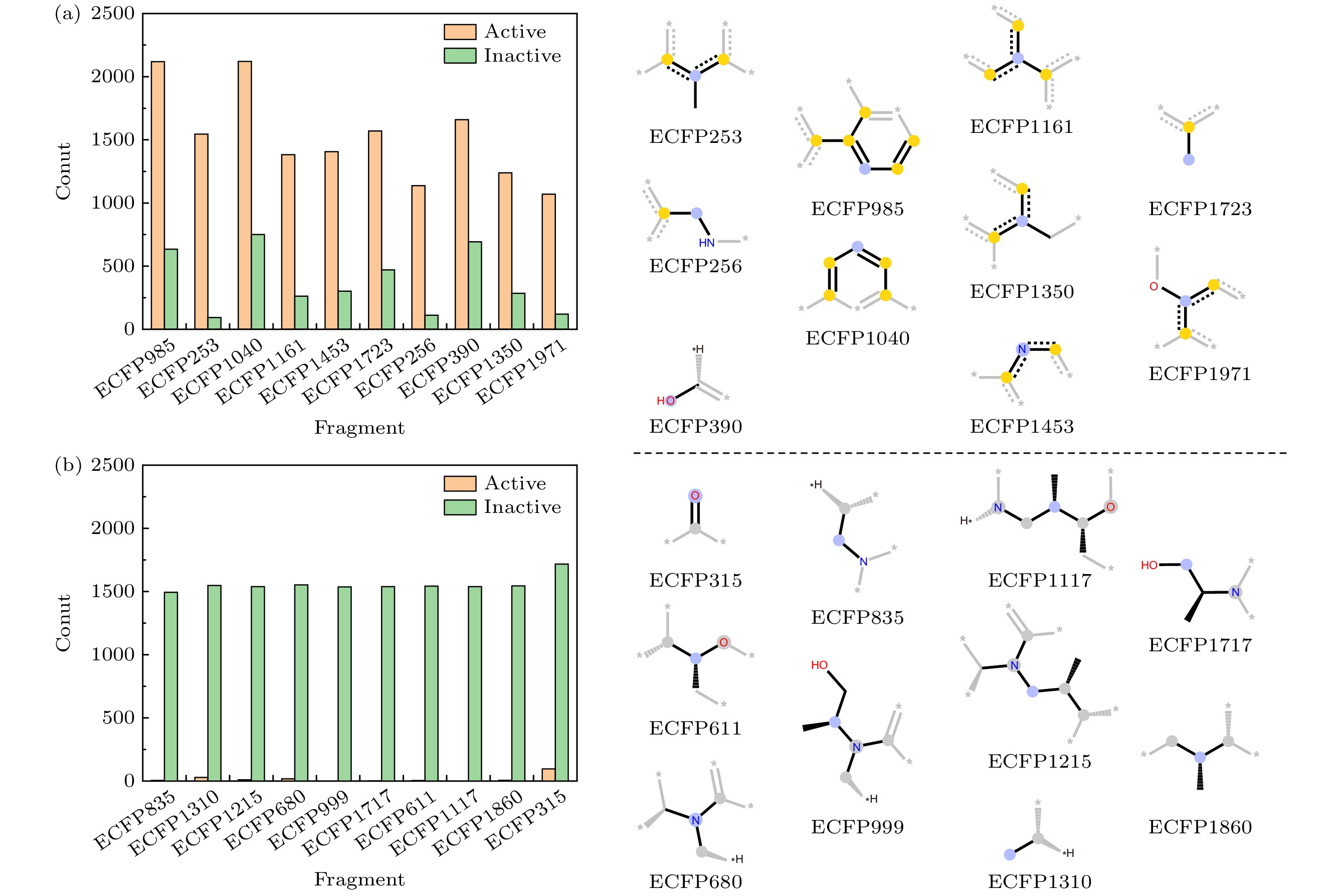
图 3 分子结构片段在两类样本间的计数差异 (a)活性化合物计数占优的结构片段; (b)非活性化合物计数占优的结构片段. 结构片段的中心原子以蓝色突出显示; 芳香性原子被着色为黄色, 而脂肪烃链原子则被着色为灰色
Fig. 3. Count difference of fragments of structures between active and inactive compounds: (a) Fragments in active compounds with a proportion higher than inactive compounds; (b) fragments in active compounds with a proportion lower than inactive compounds. The center atoms of substructure fragments are highlighted in blue; aromaticity atoms are colored in yellow and aliphatic hydrocarbon atoms are colored in gray.
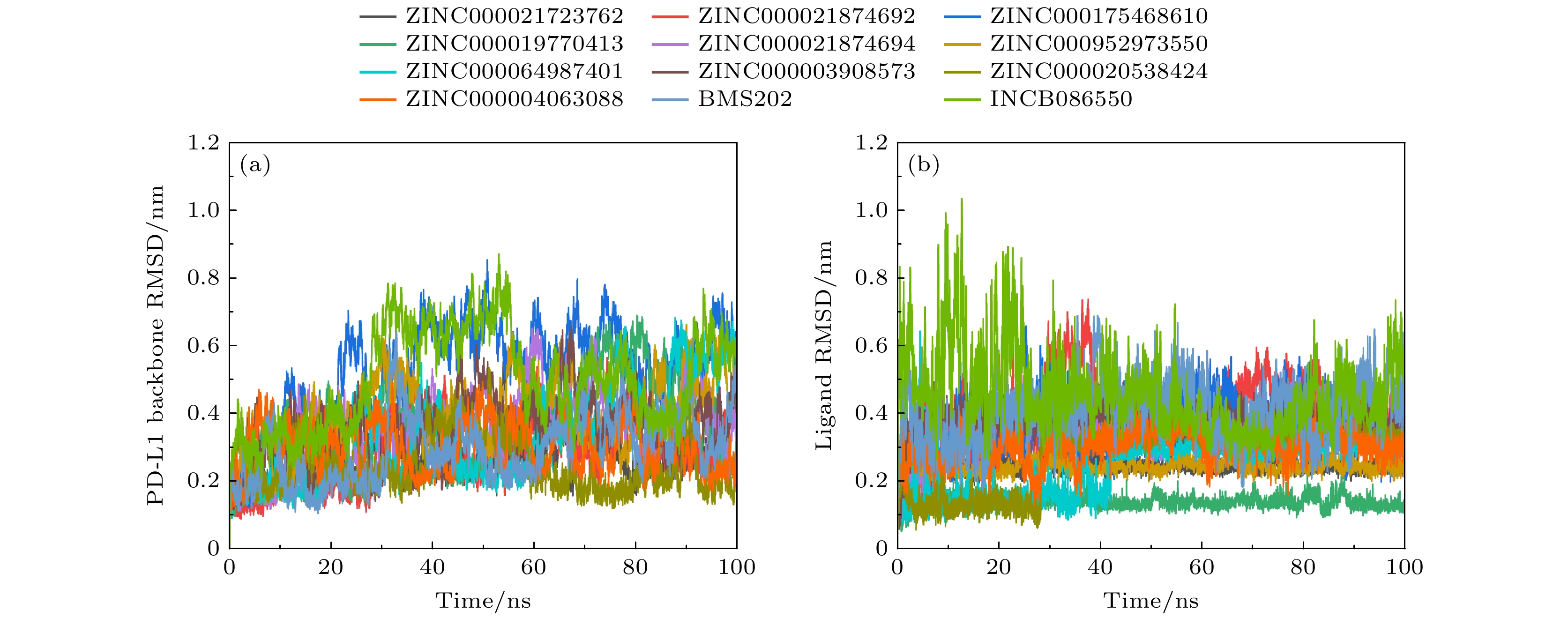
图 8 配体与PD-L1结合模式和关键残基 (a) ZINC000021723762; (b) ZINC000021874692; (c) ZINC000175468610; (d) ZINC000019770413; (e) ZINC000021874694; (f) ZINC000952973550; (g) ZINC000064987401; (h) ZINC000003908573; (i) ZINC000020538424; (j) ZINC000004063088; (k) BMS202; (l) INCB086550
Fig. 8. Ligand binding mode with PD-L1 and key residues: (a) ZINC000021723762; (b) ZINC000021874692; (c) ZINC000175468610; (d) ZINC000019770413; (e) ZINC000021874694; (f) ZINC000952973550; (g) ZINC000064987401; (h) ZINC000003908573; (i) ZINC000020538424; (j) ZINC000004063088; (k) BMS202; (l) NCB086550.
表 1 最低对接评分的10种候选化合物及BMS202和INCB086550的对接结果
Table 1. Docking results for the top 10 hits with BMS202 and INCB086550.
序号 化合物 化学结构式 对接分数/(kcal·mol–1) Hit 1 ZINC000021723762 
–11.8 Hit 2 ZINC000021874692 
–10.7 Hit 3 ZINC000175468610 
–10.7 Hit 4 ZINC000019770413 
–10.6 Hit 5 ZINC000021874694 
–10.6 Hit 6 ZINC000952973550 
–10.5 Hit 7 ZINC000064987401 
–10.5 Hit 8 ZINC000003908573 
–10.4 Hit 9 ZINC000020538424 
–10.4 Hit 10 ZINC000004063088 
–10.3 Ctr 1 BMS202 
–11.1 Ctr 2 INCB086550 
–10.0 表 2 结合自由能计算结果
Table 2. Results of MMPBSA.
化合物 $ {E}_{{\mathrm{v}}{\mathrm{d}}{\mathrm{w}}} $/(kcal·mol–1) $ {E}_{{\mathrm{e}}{\mathrm{l}}{\mathrm{e}}} $/(kcal·mol–1) $ {E}_{{\mathrm{P}}{\mathrm{B}}} $/(kcal·mol–1) $ {E}_{{\mathrm{S}}{\mathrm{A}}} $/(kcal·mol–1) $ \Delta G $/(kcal·mol–1) ZINC000021723762 –58.86$ \pm 0.12 $ –8.76$ \pm $0.10 35.53$ \pm $0.09 –4.03$ \pm $0.00 –36.12$ \pm $0.12 ZINC000021874692 –50.42$ \pm $0.11 –6.50$ \pm $0.14 35.65$ \pm $0.16 –4.50$ \pm $0.01 –25.78$ \pm 0.12 $ ZINC000175468610 –42.46$ \pm 0.09 $ –14.34$ \pm 0.08 $ 36.20$ \pm 0.13 $ –4.02$ \pm $0.00 –24.61$ \pm 0.10 $ ZINC000019770413 –47.31$ \pm 0.08 $ –3.45$ \pm 0.08 $ 26.32$ \pm 0.07 $ –3.48$ \pm $0.00 –27.91$ \pm 0.08 $ ZINC000021874694 –55.17$ \pm 0.09 $ –17.87$ \pm 0.13 $ 45.67$ \pm 0.14 $ –4.59$ \pm $0.00 –31.97$ \pm 0.12 $ ZINC000952973550 –55.82$ \pm 0.08 $ –13.47$ \pm 0.11 $ 45.83$ \pm 0.11 $ –4.19$ \pm $0.00 –27.65$ \pm 0.11 $ ZINC000064987401 –48.50$ \pm 0.08 $ –14.71$ \pm 0.12 $ 38.59$ \pm 0.11 $ –4.06$ \pm $0.00 –28.68$ \pm 0.12 $ ZINC000003908573 –44.75$ \pm 0.07 $ 4.59$ \pm 0.11 $ 20.20$ \pm 0.13 $ –3.74$ \pm $0.00 –23.70$ \pm 0.12 $ ZINC000020538424 –49.50$ \pm 0.09 $ –8.50$ \pm 0.13 $ 32.61$ \pm 0.16 $ –4.17$ \pm $0.01 –29.57$ \pm 0.10 $ ZINC000004063088 –64.14$ \pm 0.09 $ –5.38$ \pm 0.11 $ 36.75$ \pm 0.10 $ –4.21$ \pm $0.00 –36.98$ \pm 0.10 $ BMS202 –40.23$ \pm 0.08 $ –3.57$ \pm 0.07 $ 22.35$ \pm 0.10 $ –4.31$ \pm $0.01 –25.75$ \pm 0.09 $ INCB086550 –50.65$ \pm 0.14 $ –9.87$ \pm 0.17 $ 44.44$ \pm 0.27 $ –5.13$ \pm $0.02 –21.22$ \pm $0.15 -
[1] Waldman A D, Fritz J M, Lenardo M J 2020 Nat. Rev. Immunol. 20 651
 Google Scholar
Google Scholar
[2] Wherry E J, Kurachi M 2015 Nat. Rev. Immunol. 15 486
 Google Scholar
Google Scholar
[3] Zou W, Wolchok J D, Chen L 2016 Sci. Transl. Med. 8 328rv4
 Google Scholar
Google Scholar
[4] Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, Wu X, Ma J, Zhou M, Li X, Li Y, Li G, Xiong W, Guo C, Zeng Z 2019 Mol. Cancer 18 10
 Google Scholar
Google Scholar
[5] Topalian S L, Drake C G, Pardoll D M 2015 Cancer Cell 27 450
 Google Scholar
Google Scholar
[6] Postow M A, Callahan M K, Wolchok J D 2015 J. Clin. Oncol. 33 1974
 Google Scholar
Google Scholar
[7] Tang Q, Chen Y, Li X, Long S, Shi Y, Yu Y, Wu W, Han L, Wang S 2022 Front. Immunol. 13 964442
 Google Scholar
Google Scholar
[8] Ai L, Xu A, Xu J 2020 Adv. Exp. Med. Biol. 1248 33
 Google Scholar
Google Scholar
[9] Dermani F K, Samadi P, Rahmani G, Kohlan A K, Najafi R 2019 J. Cell. Physiol. 234 1313
 Google Scholar
Google Scholar
[10] Parry R V, Chemnitz J M, Frauwirth K A, Lanfranco A R, Braunstein I, Kobayashi S V, Linsley P S, Thompson C B, Riley J L 2005 Mol. Cell. Biol. 25 9543
 Google Scholar
Google Scholar
[11] Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis V A 2012 Sci. Signal. 5 ra46
 Google Scholar
Google Scholar
[12] Hui E, Cheung J, Zhu J, Su X, Taylor M J, Wallweber H A, Sasmal D K, Huang J, Kim J M, Mellman I, Vale R D 2017 Science 355 1428
 Google Scholar
Google Scholar
[13] Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N 2002 Proc. Natl. Acad. Sci. U.S.A. 99 12293
 Google Scholar
Google Scholar
[14] Iwai Y, Terawaki S, Honjo T 2005 Int. Immunol. 17 133
 Google Scholar
Google Scholar
[15] Gong J, Chehrazi-Raffle A, Reddi S, Salgia R 2018 J. Immunother. Cancer 6 8
 Google Scholar
Google Scholar
[16] Akinleye A, Rasool Z 2019 J. Hematol. Oncol. 12 92
 Google Scholar
Google Scholar
[17] Shiravand Y, Khodadadi F, Kashani S M A, Hosseini-Fard S R, Hosseini S, Sadeghirad H, Ladwa R, O'Byrne K, Kulasinghe A 2022 Curr. Oncol. 29 3044
 Google Scholar
Google Scholar
[18] Zhang J, Zhang Y, Qu B, Yang H, Hu S, Dong X 2021 Eur. J. Med. Chem. 218 113356
 Google Scholar
Google Scholar
[19] Johnson D B, Nebhan C A, Moslehi J J, Balko J M 2022 Nat. Rev. Clin. Oncol. 19 254
 Google Scholar
Google Scholar
[20] Mould D R, Meibohm B 2016 BioDrugs 30 275
 Google Scholar
Google Scholar
[21] Wu Q, Jiang L, Li S C, He Q J, Yang B, Cao J 2021 Acta Pharmacol. Sin. 42 1
 Google Scholar
Google Scholar
[22] Zhan M M, Hu X Q, Liu X X, Ruan B F, Xu J, Liao C 2016 Drug Discov. Today 21 1027
 Google Scholar
Google Scholar
[23] Liu C, Seeram N P, Ma H 2021 Cancer Cell Int. 21 239
 Google Scholar
Google Scholar
[24] Chupak L S, Zheng X 2015 WO Patent 2015034820 A1
[25] Zak K M, Grudnik P, Guzik K, Zieba B J, Musielak B, Dömling A, Dubin G, Holak T A 2016 Oncotarget 7 30323
 Google Scholar
Google Scholar
[26] Koblish H K, Wu L, Wang L S, Liu P C C, Wynn R, Rios-Doria J, Spitz S, Liu H, Volgina A, Zolotarjova N, Kapilashrami K, Behshad E, Covington M, Yang Y O, Li J, Diamond S, Soloviev M, O'Hayer K, Rubin S, Kanellopoulou C, Yang G, Rupar M, DiMatteo D, Lin L, Stevens C, Zhang Y, Thekkat P, Geschwindt R, Marando C, Yeleswaram S, Jackson J, Scherle P, Huber R, Yao W, Hollis G 2022 Cancer Discov. 12 1482
 Google Scholar
Google Scholar
[27] Jiang J, Zou X, Liu Y, Liu X, Dong K, Yao X, Feng Z, Chen X, Sheng L, Li Y 2021 Front. Pharmacol. 12 677120
 Google Scholar
Google Scholar
[28] Wang Y, Liu X, Zou X, Wang S, Luo L, Liu Y, Dong K, Yao X, Li Y, Chen X, Sheng L 2021 Pharmaceutics 13 598
 Google Scholar
Google Scholar
[29] Sobral P S, Luz V C C, Almeida J, Videira P A, Pereira F 2023 Int. J. Mol. Sci. 24 5908
 Google Scholar
Google Scholar
[30] Luo L, Zhong A, Wang Q, Zheng T 2021 Mar. Drugs 20 29
 Google Scholar
Google Scholar
[31] Acúrcio R C, Pozzi S, Carreira B, Pojo M, Gómez-Cebrián N, Casimiro S, Fernandes A, Barateiro A, Farricha V, Brito J, Leandro A P, Salvador J A R, Graça L, Puchades-Carrasco L, Costa L, Satchi-Fainaro R, Guedes R C, Florindo H F 2022 J. Immunother. Cancer 10 e004695
 Google Scholar
Google Scholar
[32] Lung J, Hung M S, Lin Y C, Hung C H, Chen C C, Lee K D, Tsai Y H 2020 Molecules 25 5293
 Google Scholar
Google Scholar
[33] Guo Y, Liang J, Liu B, Jin Y 2021 Int. J. Mol. Sci. 22 10924
 Google Scholar
Google Scholar
[34] Sterling T, Irwin J J 2015 J. Chem. Inf. Model. 55 2324
 Google Scholar
Google Scholar
[35] Bemis G W, Murcko M A 1996 J. Med. Chem. 39 2887
 Google Scholar
Google Scholar
[36] Rogers D, Hahn M 2010 J. Chem. Inf. Model. 50 742
 Google Scholar
Google Scholar
[37] Yap C W 2011 J. Comput. Chem. 32 1466
 Google Scholar
Google Scholar
[38] Durant J L, Leland B A, Henry D R, Nourse J G 2002 J. Chem. Inf. Comput. Sci. 42 1273
 Google Scholar
Google Scholar
[39] Moriwaki H, Tian Y S, Kawashita N, Takagi T 2018 J. Cheminform. 10 4
 Google Scholar
Google Scholar
[40] Bickerton G R, Paolini G V, Besnard J, Muresan S, Hopkins A L 2012 Nat. Chem. 4 90
 Google Scholar
Google Scholar
[41] Kosugi T, Ohue M 2021 Int. J. Mol. Sci. 22 10925
 Google Scholar
Google Scholar
[42] Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D 2021 Nucleic Acids Res. 49 W5
 Google Scholar
Google Scholar
[43] Jing T, Zhang Z, Kang Z, Mo J, Yue X, Lin Z, Fu X, Liu C, Ma H, Zhang X, Hu W 2023 J. Med. Chem. 66 6811
 Google Scholar
Google Scholar
[44] Qin M, Meng Y, Yang H, Liu L, Zhang H, Wang S, Liu C, Wu X, Wu D, Tian Y, Hou Y, Zhao Y, Liu Y, Xu C, Wang L 2021 J. Med. Chem. 64 5519
 Google Scholar
Google Scholar
[45] Trott O, Olson A J 2010 J. Comput. Chem. 31 455
 Google Scholar
Google Scholar
[46] Laskowski R A, Swindells M B 2011 J. Chem. Inf. Model. 51 2778
 Google Scholar
Google Scholar
[47] Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell Jr. A D 2010 J. Comput. Chem. 31 671
 Google Scholar
Google Scholar
[48] Yu W, He X, Vanommeslaeghe K, MacKerell Jr. A D 2012 J. Comput. Chem. 33 2451
 Google Scholar
Google Scholar
[49] Vanommeslaeghe K, MacKerell Jr. A D 2012 J. Chem. Inf. Model. 52 3144
 Google Scholar
Google Scholar
[50] Vanommeslaeghe K, Raman E P, MacKerell Jr. A D 2012 J. Chem. Inf. Model. 52 3155
 Google Scholar
Google Scholar
[51] Abraham M J, Murtola T, Schulz R, Páll S, Smith J C, Hess B, Lindahl E 2015 SoftwareX 1 19
 Google Scholar
Google Scholar
[52] Parrinello M, Rahman A 1981 J. Appl. Phys. 52 7182
 Google Scholar
Google Scholar
[53] Nosé S 1984 Mol. Phys. 52 255
 Google Scholar
Google Scholar
[54] Hoover W G 1985 Phys. Rev. A 31 1695
 Google Scholar
Google Scholar
[55] Essmann U, Perera L, Berkowitz M L, Darden T, Lee H, Pedersen L G 1995 J. Chem. Phys. 103 8577
 Google Scholar
Google Scholar
[56] Hess B, Bekker H, Berendsen H J C, Fraaije J G E M 1997 J. Comput. Chem. 18 1463
 Google Scholar
Google Scholar
[57] Valdés-Tresanco M S, Valdés-Tresanco M E, Valiente P A, Moreno E 2021 J. Chem. Theory Comput. 17 6281
 Google Scholar
Google Scholar
[58] Chupak L S, Ding M, Martin S W, Zheng X, Hewawasam P, Connolly T P, Xu N, Yeung K S, Zhu J, Langley D R, Tenney D J, Scola P M 2015 WO Patent 2015160641 A3
[59] Wang T, Cai S, Wang M, Zhang W, Zhang K, Chen D, Li Z, Jiang S 2021 J. Med. Chem. 64 7390
 Google Scholar
Google Scholar
[60] Lu L, Qi Z, Wang T, Zhang X, Zhang K, Wang K, Cheng Y, Xiao Y, Li Z, Jiang S 2022 ACS Med. Chem. Lett. 13 586
 Google Scholar
Google Scholar
[61] Dai X, Wang K, Chen H, Huang X, Feng Z 2021 Bioorg. Chem. 114 105034
 Google Scholar
Google Scholar
[62] Le Biannic R, Magnez R, Klupsch F, Leleu-Chavain N, Thiroux B, Tardy M, El Bouazzati H, Dezitter X, Renault N, Vergoten G, Bailly C, Quesnel B, Thuru X, Millet R 2022 Eur. J. Med. Chem. 236 114343
 Google Scholar
Google Scholar
[63] Russomanno P, Assoni G, Amato J, D'Amore V M, Scaglia R, Brancaccio D, Pedrini M, Polcaro G, La Pietra V, Orlando P, Falzoni M, Cerofolini L, Giuntini S, Fragai M, Pagano B, Donati G, Novellino E, Quintavalle C, Condorelli G, Sabbatino F, Seneci P, Arosio D, Pepe S, Marinelli L 2021 J. Med. Chem. 64 16020
 Google Scholar
Google Scholar
[64] Liu L, Yao Z, Wang S, Xie T, Wu G, Zhang H, Zhang P, Wu Y, Yuan H, Sun H 2021 J. Med. Chem. 64 8391
 Google Scholar
Google Scholar
[65] Cheng B, Ren Y, Cao H, Chen J 2020 Eur. J. Med. Chem. 199 112377
 Google Scholar
Google Scholar
[66] Lipinski C A 2004 Drug Discov. Today Technol. 1 337
 Google Scholar
Google Scholar
[67] Liang J, Wang B, Yang Y, Liu B, Jin Y 2023 Int. J. Mol. Sci. 24 1280
 Google Scholar
Google Scholar
计量
- 文章访问数: 5835
- PDF下载量: 166
- 被引次数: 0















 下载:
下载: