-
蛋白质分子的“液-液相分离”是近几年生物物理学领域迅速发展起来的研究热点. 蛋白质相分离在一系列生物学过程中发挥着重要的作用. 蛋白质分子序列和构象的多样性和复杂性, 给蛋白质分子的理论研究、计算机模拟和实验研究都带来了巨大的挑战. 当前, 多尺度理论模型和多分辨率计算方法被广泛地用于蛋白质分子的“液-液相分离”的研究中. 本文将对蛋白质分子“液-液相分离”的理论基础和计算机模拟方法进行简要的综述, 并对这些理论和方法未来的发展趋势进行了初步的探讨和展望. 期望为进一步研究蛋白质“液-液相分离”的物理化学机制和过程提供理论基础和方法借鉴.Liquid-liquid phase separation (LLPS) of proteins is an emerging field in the research of biophysics. Many intrinsically disordered proteins (IDPs) are known to have the ability to assemble via LLPS and to organize into protein-rich and dilute phases both in vivo and in vitro. Such a kind of phase separation of proteins plays an important role in a wide range of cellular processes, such as the formation of membraneless organelles (MLOs), signaling transduction, intracellular organization, chromatin organization, etc. In recent years, there appeared a great number of theoretical analysis, computational simulation and experimental research focusing on the physical principles of LLPS. In this article, the theoretical and computational simulation methods for the LLPS are briefly reviewed. To elucidate the physical principle of LLPS and to understand the phase behaviors of the proteins, biophysicists have introduced the concepts and theories from statistical mechanics and polymer sciences. Flory-Huggins theory and its extensions, such as mean-field model, random phase approximation (RPA) and field theory simulations, can conduce to understanding the phase diagram of the LLPS. To reveal the hidden principles in the sequence-dependent phase behaviors of different biomolecular condensates, different simulation methods including lattice models, off-lattice coarse-grained models, and all-atom simulations are introduced to perform computer simulations. By reducing the conformational space of the proteins, lattice models can capture the key points in LLPS and simplify the computations. In the off-lattice models, a polypeptide can be coarse-grained as connected particles representing repeated short peptide fragments. All-atom simulations can describe the structure of proteins at a higher resolution but consume higher computation-power. Multi-scale simulation may provide the key to understanding LLPS at both high computational efficiency and high accuracy. With these methods, we can elucidate the sequence-dependent phase behaviors of proteins at different resolutions. To sum up, it is necessary to choose the appropriate method to model LLPS processes according to the interactions within the molecules and the specific phase behaviors of the system. The simulations of LLPS can facilitate the comprehensive understanding of the key features which regulate the membraneless compartmentalization in cell biology and shed light on the design of artificial cells and the control of neurodegeneration.
-
Keywords:
- liquid-liquid phase separation /
- intrinsically disordered proteins /
- multivalent interaction /
- multi-scale simulation
[1] McCarty J, Delaney K T, Danielsen S P, Fredrickson G H, Shea J E 2019 J. Phys. Chem. Lett. 10 1644
 Google Scholar
Google Scholar
[2] Lin Y H, Song J H, Forman-Kay J D, Chan H S 2017 J. Mol. Liq. 228 176
 Google Scholar
Google Scholar
[3] Lin Y H, Song J H, Forman-Kay J D, Chan H S 2019 J. Mol. Liq. 273 676
 Google Scholar
Google Scholar
[4] Dignon G L, Zheng W W, Kim Y C, Mittal J, Best R B 2018 Biophys. J. 114 431a
 Google Scholar
Google Scholar
[5] Dignon G L, Zheng W W, Best R B, Mittal J 2017 Biophys. J. 112 200a
[6] Choi J M, Mitrea D M, Stanley C B, Ruff K M, Holehouse A S, Kriwacki R W, Pappu R V 2019 Biophys. J. 116 349a
 Google Scholar
Google Scholar
[7] Chin A F, Hilser V J, Zheng Y X 2019 Biophys. J. 116 179a
 Google Scholar
Google Scholar
[8] 周丰茂, 孙东科, 朱鸣芳 2010 59 3394
 Google Scholar
Google Scholar
Zhou F M, Sun D K, Zhu M F 2010 Acta Phys. Sin. 59 3394
 Google Scholar
Google Scholar
[9] 张长胜, 来鲁华 2010 物理化学学报 36 1907053
 Google Scholar
Google Scholar
Zhang C S, Lai L H 2010 Acta Phys. Chim. Sin. 36 1907053
 Google Scholar
Google Scholar
[10] Burke K A, Janke A M, Rhine C L, Fawzi N L 2015 Mol. Cell 60 231
 Google Scholar
Google Scholar
[11] Brangwynne C P, Tompa P, Pappu R V 2015 Nat. Phys. 11 899
 Google Scholar
Google Scholar
[12] Uversky V N 2017 Curr. Opin. Struct. Biol. 44 18
 Google Scholar
Google Scholar
[13] Feric M, Vaidya N, Harmon T S, Mitrea D M, Zhu L, Richardson T M, Kriwacki R W, Pappu R V, Brangwynne C P 2016 Cell 165 1686
 Google Scholar
Google Scholar
[14] Falahati H, Wieschaus E 2017 Proc. Natl. Acad. Sci. U.S.A. 114 1335
 Google Scholar
Google Scholar
[15] Sabari B R, Dall'Agnese A, Boija A, Klein I A, Coffey E L, Shrinivas K, Abraham B J, Hannett N M, Zamudio A V, Manteiga J C, Li C H, Guo Y E, Day D S, Schuijers J, Vasile E, Malik S, Hnisz D, Lee T I, Cisse, I I, Roeder R G, Sharp P A, Chakraborty A K, Young R A 2018 Science 361 eaar3958
 Google Scholar
Google Scholar
[16] Das R K, Pappu R V 2013 Proc. Natl. Acad. Sci. U.S.A. 110 13392
 Google Scholar
Google Scholar
[17] Dignon G L, Zheng W W, Best R B, Kim Y C, Mittal J 2018 Proc. Natl. Acad. Sci. U.S.A. 115 9929
 Google Scholar
Google Scholar
[18] Harmon T S, Holehouse A S, Rosen M K, Pappu R V 2017 ELIFE 6 e30294
 Google Scholar
Google Scholar
[19] Kang W B, He C, Liu Z X, Wang J, Wang W 2019 J. Biomol. Struct. Dyn. 37 1956
 Google Scholar
Google Scholar
[20] Lin Y, Currie S L, Rosen M K 2017 J. Biol. Chem. 292 19110
 Google Scholar
Google Scholar
[21] Lin Y H, Brady J P, Forman-Kay J D, Chan H S 2017 New J. Phys. 19 115003
 Google Scholar
Google Scholar
[22] Schuster B S, Dignon G, Jahnke C, Good M C, Hammer D A, Mittal J 2019 Biophys. J. 116 453a
 Google Scholar
Google Scholar
[23] Smith J, Calidas D, Schmidt H, Lu T, Seydoux G 2016 ELIFE 5 e21337
 Google Scholar
Google Scholar
[24] Uebel C J, Anderson D C, Mandarino L M, Manage K I, Aynaszyan S, Phillips C M 2018 Plos Genet. 14 e1007542
 Google Scholar
Google Scholar
[25] 康文斌, 王骏, 王炜 2018 67 058701
 Google Scholar
Google Scholar
Kang W B, Wang J, Wang W 2018 Acta Phys. Sin. 67 058701
 Google Scholar
Google Scholar
[26] Pak C W, Kosno M, Holehouse A S, Padrick S B, Mittal A, A R, Yunus A A, Liu D R, Pappu R V, Rosen M K 2016 Mol. Cell 63 72
 Google Scholar
Google Scholar
[27] Gates Z P, Baxa M C, Yu W, Riback J A, Li H, Roux B, Kent S B, Sosnick T R 2017 Proc. Natl. Acad. Sci. U.S.A. 114 2241
 Google Scholar
Google Scholar
[28] Riback J A, Katanski C D, Kear-Scott J L, Pilipenko E V, Rojek A E, Sosnick T R, Drummond D A 2017 Cell 168 1028
 Google Scholar
Google Scholar
[29] Das R K, Ruff K M, Pappu R V 2015 Curr. Opin. Struct. Biol. 32 102
 Google Scholar
Google Scholar
[30] Vitalis A, Wang X, Pappu R V 2007 Biophys. J. 93 1923
 Google Scholar
Google Scholar
[31] Papoian G A 2008 Proc. Natl. Acad. Sci. U. S. A. 105 14237
 Google Scholar
Google Scholar
[32] Levine Z A, Shea J E 2017 Curr. Opin. Struct. Biol. 43 95
 Google Scholar
Google Scholar
[33] Dignon G L, Zheng W, Kim Y C, Mittal J 2019 ACS Central Sci. 5 821
 Google Scholar
Google Scholar
[34] Murthy A C, Dignon G L, Kan Y, Zerze G H, Parekh S H, Mittal J, Fawzi N L 2019 Nat. Struct. Mol. Biol. 26 637
 Google Scholar
Google Scholar
[35] Ruff K M, Pappu R V, Holehouse A S 2019 Curr. Opin. Struct. Biol. 56 1
 Google Scholar
Google Scholar
[36] Monahan Z, Ryan V H, Janke A M, Burke K A, Rhoads S N, Zerze G H, O'Meally R, Dignon G L, Conicella A E, Zheng W W, Best R B, Cole R N, Mittal J, Shewmaker F, Fawzi N L 2017 EMBO J. 36 2951
 Google Scholar
Google Scholar
[37] Best R B 2017 Curr. Opin. Struct. Biol. 42 147
 Google Scholar
Google Scholar
[38] Robustelli P, Piana S, Shaw D E 2018 Proc. Natl. Acad. Sci. U.S.A. 115 E4758
 Google Scholar
Google Scholar
[39] van L R, Buljan M, Lang B, Weatheritt R J, Daughdrill G W, Dunker A K, Fuxreiter M, Gough J, Gsponer J, Jones D T, Kim P M, Kriwacki R W, Oldfield C J, Pappu R V, Tompa P, Uversky V N, Wright P E, Babu M M 2014 Chem. Rev. 114 6589
 Google Scholar
Google Scholar
[40] Mittag T, Forman-Kay J D 2007 Curr. Opin. Struct. Biol. 17 3
 Google Scholar
Google Scholar
[41] Flory P J 1942 J. Chem. Phys. 10 51
 Google Scholar
Google Scholar
[42] Huggins M L 1942 J. Chem. Phys. 46 151
 Google Scholar
Google Scholar
[43] Samanta H S, Zhuravlev P I, Hinczewski M, Hori N, Chakrabarti S, Thirumalai D 2017 Soft Matter 13 3622
 Google Scholar
Google Scholar
[44] Zhou H X, Nguemaha V, Mazarakos K, Qin S B 2018 Trends Biochem. Sci. 43 499
 Google Scholar
Google Scholar
[45] Lin Y H, Forman-Kay J D, Chan H S 2016 Phys. Rev. Lett. 117 178101
 Google Scholar
Google Scholar
[46] Liu Y X, Zhang H D, Tong C H, Yang Y L 2011 Macromolecules 44 8261
 Google Scholar
Google Scholar
[47] Speck T, Menzel A M, Bialke J, Lowen H 2015 J. Chem. Phys. 142 224109
 Google Scholar
Google Scholar
[48] Burke M G, Woscholski R, Yaliraki S N 2003 Proc. Natl. Acad. Sci. U.S.A. 100 13928
 Google Scholar
Google Scholar
[49] Ruff K M, Khan S J, Pappu R V 2014 Biophys. J. 107 1226
 Google Scholar
Google Scholar
[50] Zuccato C, Valenza M, Cattaneo E 2010 Physiol. Rev. 90 905
 Google Scholar
Google Scholar
[51] Condon J E, Martin T B, Jayaraman A 2017 Soft Matter 13 2907
 Google Scholar
Google Scholar
[52] Dignon G L, Zheng W, Kim Y C, Best R B, Mittal J 2018 PLOS Comput. Biol. 14 e1005941
 Google Scholar
Google Scholar
[53] Das S, Eisen A, Lin Y H, Chan H S 2018 J. Phys. Chem. B 122 5418
 Google Scholar
Google Scholar
[54] Kapcha L H, Rossky P J 2014 J. Mol. Boil. 426 484
 Google Scholar
Google Scholar
[55] Ashbaugh H S, Hatch H W 2008 J. Am. Chem. Soc. 130 9536
 Google Scholar
Google Scholar
[56] Ghavami A, Veenhoff L M, van der Giessen E, Onck P R 2014 Biophys. J. 107 1393
 Google Scholar
Google Scholar
[57] Borgia A, Borgia M B, Bugge K, Kissling V M, Heidarsson P O, Fernandes C B, Sottini A, Soranno A, Buholzer K J, Nettels D, Kragelund B B, Best R B, Schuler B 2018 Nature 555 61
 Google Scholar
Google Scholar
[58] Wang J, Choi J M, Holehouse A S, Lee H O, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu R V, Alberti S, Hyman A A 2018 Cell 174 688
 Google Scholar
Google Scholar
[59] Song J, Gomes G N, Shi T, Gradinaru C C, Chan H S 2017 Biophys. J. 113 1012
 Google Scholar
Google Scholar
[60] Noid W G 2013 J. Chem. Phys. 139 090901
 Google Scholar
Google Scholar
[61] Voegler Smith A, Hall C K 2001 Proteins 44 344
 Google Scholar
Google Scholar
[62] Marchut A J, Hall C K 2006 Biophys. J. 90 4574
 Google Scholar
Google Scholar
[63] Marchut A J, Hall C K 2007 Proteins 66 96
 Google Scholar
Google Scholar
[64] Nguyen H D, Hall C K 2006 J. Am. Chem. Soc. 128 1890
 Google Scholar
Google Scholar
[65] Cheon M, Chang I, Hall C K 2010 Proteins 78 2950
 Google Scholar
Google Scholar
[66] Yeo J J, Huang W W, Tarakanova A, Zhang Y W, Kaplan D L, Buehler M J 2018 J. Mater. Chem. B 6 3727
 Google Scholar
Google Scholar
[67] Bereau T, Deserno M 2009 J. Chem. Phys. 130 235106
 Google Scholar
Google Scholar
[68] Rutter G O, Brown A H, Quigley D, Walsh T R, Allen M P 2015 Phys. Chem. Chem. Phys. 17 31741
 Google Scholar
Google Scholar
[69] Chen M C, Wolynes P G 2017 Proc. Natl. Acad. Sci. U.S.A. 114 4406
 Google Scholar
Google Scholar
[70] Seo M, Rauscher S, Pomes R, Tieleman D P 2012 J. Chem. Theory Comput. 8 1774
 Google Scholar
Google Scholar
[71] Miao L, Schulten K 2009 Structure 17 449
 Google Scholar
Google Scholar
[72] Monticelli L, Kandasamy S K, Periole X, Larson R G, Tieleman D P, Marrink S J 2008 J. Chem. Theory Comput. 4 819
 Google Scholar
Google Scholar
[73] Marrink S J, Risselada H J, Yefimov S, Tieleman D P, Vries A H 2007 J. Phys. Chem. B 111 7812
 Google Scholar
Google Scholar
[74] Lu L Y, Dama J F, Voth G A 2013 J. Chem. Phys. 139 121906
 Google Scholar
Google Scholar
[75] Hills R D, Lu L, Voth G A 2010 PLOS Comput. Biol. 6 e1000827
 Google Scholar
Google Scholar
[76] Ando D, Zandi R, Kim Y W, Colvin M, Rexach M, Gopinathan A J 2014 Biophys. J. 106 1997
 Google Scholar
Google Scholar
[77] Kim Y C, Hummer G 2008 J. Mol. Biol. 375 1416
 Google Scholar
Google Scholar
[78] Ryan V H, Dignon G L, Zerze G H, Chabata C V, Silva R, Conicella A E, Amaya J, Burke K A, Mittal J, Fawzi N L 2018 Mol. Cell 69 465
 Google Scholar
Google Scholar
[79] Choi J-M, Dar F, Pappu R V 2019 PLOS Comput. Biol. 15 e1007028
 Google Scholar
Google Scholar
[80] Tran H T, Mao A, Pappu R V 2008 J. Am. Chem. Soc. 130 7380
 Google Scholar
Google Scholar
[81] Holehouse A S, Garai K, Lyle N, Vitalis A, Pappu R V 2015 J. Am. Chem. Soc. 137 2984
 Google Scholar
Google Scholar
[82] Asthagiri D, Karandur D, Tomar D S, Pettitt B M 2017 J. Phys. Chem. B 121 8078
 Google Scholar
Google Scholar
[83] Karandur D, Wong K Y, Pettitt B M 2014 J. Phys. Chem. B 118 9565
 Google Scholar
Google Scholar
[84] Vitalis A, Wang X, Pappu R V 2008 J. Mol. Biol. 384 279
 Google Scholar
Google Scholar
[85] Vitalis A, Lyle N, Pappu R V 2009 Biophys. J. 97 303
 Google Scholar
Google Scholar
[86] Kang H, Vazquez F X, Zhang L, Das P, Toledo-Sherman L, Luan B, Levitt M, Zhou R 2017 J. Am. Chem. Soc. 139 8820
 Google Scholar
Google Scholar
[87] Daggett V, Kollman P A, Kuntz I D 1991 Biopolymers 31 1115
 Google Scholar
Google Scholar
-
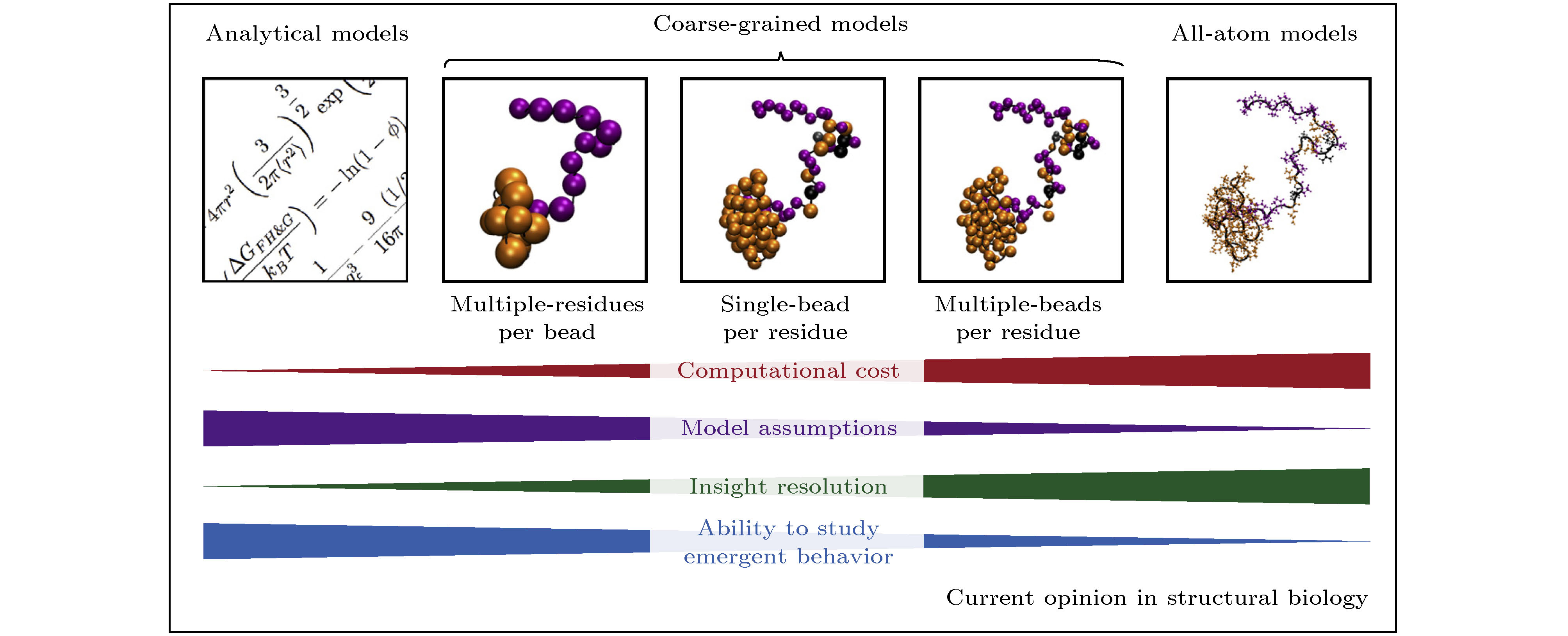
图 1 研究蛋白质“液-液相分离”的计算方法: 从左至右分别是解析模型、粗粒化模型(包括多个残基单个粒子、单个残基单个粒子、单个残基多个粒子的模型)和全原子模型, 分辨率的提升对计算资源的耗费程度急剧增高, 而模型假设数的减少会减弱研究涌现行为的能力[35]
Fig. 1. Computational approaches utilized to study protein liquid-liquid phase separation (LLPS). From left to right are the analytical model, the coarse-grained model (multiple-residues per bead, single-bead per residue and multiple-beads per residue coarse-grained models), and all-atom model. High-resolution descriptions increase the computational cost of the simulations. All-atom model is often impractical for the study of emergent behavior of LLPS[35].
-
[1] McCarty J, Delaney K T, Danielsen S P, Fredrickson G H, Shea J E 2019 J. Phys. Chem. Lett. 10 1644
 Google Scholar
Google Scholar
[2] Lin Y H, Song J H, Forman-Kay J D, Chan H S 2017 J. Mol. Liq. 228 176
 Google Scholar
Google Scholar
[3] Lin Y H, Song J H, Forman-Kay J D, Chan H S 2019 J. Mol. Liq. 273 676
 Google Scholar
Google Scholar
[4] Dignon G L, Zheng W W, Kim Y C, Mittal J, Best R B 2018 Biophys. J. 114 431a
 Google Scholar
Google Scholar
[5] Dignon G L, Zheng W W, Best R B, Mittal J 2017 Biophys. J. 112 200a
[6] Choi J M, Mitrea D M, Stanley C B, Ruff K M, Holehouse A S, Kriwacki R W, Pappu R V 2019 Biophys. J. 116 349a
 Google Scholar
Google Scholar
[7] Chin A F, Hilser V J, Zheng Y X 2019 Biophys. J. 116 179a
 Google Scholar
Google Scholar
[8] 周丰茂, 孙东科, 朱鸣芳 2010 59 3394
 Google Scholar
Google Scholar
Zhou F M, Sun D K, Zhu M F 2010 Acta Phys. Sin. 59 3394
 Google Scholar
Google Scholar
[9] 张长胜, 来鲁华 2010 物理化学学报 36 1907053
 Google Scholar
Google Scholar
Zhang C S, Lai L H 2010 Acta Phys. Chim. Sin. 36 1907053
 Google Scholar
Google Scholar
[10] Burke K A, Janke A M, Rhine C L, Fawzi N L 2015 Mol. Cell 60 231
 Google Scholar
Google Scholar
[11] Brangwynne C P, Tompa P, Pappu R V 2015 Nat. Phys. 11 899
 Google Scholar
Google Scholar
[12] Uversky V N 2017 Curr. Opin. Struct. Biol. 44 18
 Google Scholar
Google Scholar
[13] Feric M, Vaidya N, Harmon T S, Mitrea D M, Zhu L, Richardson T M, Kriwacki R W, Pappu R V, Brangwynne C P 2016 Cell 165 1686
 Google Scholar
Google Scholar
[14] Falahati H, Wieschaus E 2017 Proc. Natl. Acad. Sci. U.S.A. 114 1335
 Google Scholar
Google Scholar
[15] Sabari B R, Dall'Agnese A, Boija A, Klein I A, Coffey E L, Shrinivas K, Abraham B J, Hannett N M, Zamudio A V, Manteiga J C, Li C H, Guo Y E, Day D S, Schuijers J, Vasile E, Malik S, Hnisz D, Lee T I, Cisse, I I, Roeder R G, Sharp P A, Chakraborty A K, Young R A 2018 Science 361 eaar3958
 Google Scholar
Google Scholar
[16] Das R K, Pappu R V 2013 Proc. Natl. Acad. Sci. U.S.A. 110 13392
 Google Scholar
Google Scholar
[17] Dignon G L, Zheng W W, Best R B, Kim Y C, Mittal J 2018 Proc. Natl. Acad. Sci. U.S.A. 115 9929
 Google Scholar
Google Scholar
[18] Harmon T S, Holehouse A S, Rosen M K, Pappu R V 2017 ELIFE 6 e30294
 Google Scholar
Google Scholar
[19] Kang W B, He C, Liu Z X, Wang J, Wang W 2019 J. Biomol. Struct. Dyn. 37 1956
 Google Scholar
Google Scholar
[20] Lin Y, Currie S L, Rosen M K 2017 J. Biol. Chem. 292 19110
 Google Scholar
Google Scholar
[21] Lin Y H, Brady J P, Forman-Kay J D, Chan H S 2017 New J. Phys. 19 115003
 Google Scholar
Google Scholar
[22] Schuster B S, Dignon G, Jahnke C, Good M C, Hammer D A, Mittal J 2019 Biophys. J. 116 453a
 Google Scholar
Google Scholar
[23] Smith J, Calidas D, Schmidt H, Lu T, Seydoux G 2016 ELIFE 5 e21337
 Google Scholar
Google Scholar
[24] Uebel C J, Anderson D C, Mandarino L M, Manage K I, Aynaszyan S, Phillips C M 2018 Plos Genet. 14 e1007542
 Google Scholar
Google Scholar
[25] 康文斌, 王骏, 王炜 2018 67 058701
 Google Scholar
Google Scholar
Kang W B, Wang J, Wang W 2018 Acta Phys. Sin. 67 058701
 Google Scholar
Google Scholar
[26] Pak C W, Kosno M, Holehouse A S, Padrick S B, Mittal A, A R, Yunus A A, Liu D R, Pappu R V, Rosen M K 2016 Mol. Cell 63 72
 Google Scholar
Google Scholar
[27] Gates Z P, Baxa M C, Yu W, Riback J A, Li H, Roux B, Kent S B, Sosnick T R 2017 Proc. Natl. Acad. Sci. U.S.A. 114 2241
 Google Scholar
Google Scholar
[28] Riback J A, Katanski C D, Kear-Scott J L, Pilipenko E V, Rojek A E, Sosnick T R, Drummond D A 2017 Cell 168 1028
 Google Scholar
Google Scholar
[29] Das R K, Ruff K M, Pappu R V 2015 Curr. Opin. Struct. Biol. 32 102
 Google Scholar
Google Scholar
[30] Vitalis A, Wang X, Pappu R V 2007 Biophys. J. 93 1923
 Google Scholar
Google Scholar
[31] Papoian G A 2008 Proc. Natl. Acad. Sci. U. S. A. 105 14237
 Google Scholar
Google Scholar
[32] Levine Z A, Shea J E 2017 Curr. Opin. Struct. Biol. 43 95
 Google Scholar
Google Scholar
[33] Dignon G L, Zheng W, Kim Y C, Mittal J 2019 ACS Central Sci. 5 821
 Google Scholar
Google Scholar
[34] Murthy A C, Dignon G L, Kan Y, Zerze G H, Parekh S H, Mittal J, Fawzi N L 2019 Nat. Struct. Mol. Biol. 26 637
 Google Scholar
Google Scholar
[35] Ruff K M, Pappu R V, Holehouse A S 2019 Curr. Opin. Struct. Biol. 56 1
 Google Scholar
Google Scholar
[36] Monahan Z, Ryan V H, Janke A M, Burke K A, Rhoads S N, Zerze G H, O'Meally R, Dignon G L, Conicella A E, Zheng W W, Best R B, Cole R N, Mittal J, Shewmaker F, Fawzi N L 2017 EMBO J. 36 2951
 Google Scholar
Google Scholar
[37] Best R B 2017 Curr. Opin. Struct. Biol. 42 147
 Google Scholar
Google Scholar
[38] Robustelli P, Piana S, Shaw D E 2018 Proc. Natl. Acad. Sci. U.S.A. 115 E4758
 Google Scholar
Google Scholar
[39] van L R, Buljan M, Lang B, Weatheritt R J, Daughdrill G W, Dunker A K, Fuxreiter M, Gough J, Gsponer J, Jones D T, Kim P M, Kriwacki R W, Oldfield C J, Pappu R V, Tompa P, Uversky V N, Wright P E, Babu M M 2014 Chem. Rev. 114 6589
 Google Scholar
Google Scholar
[40] Mittag T, Forman-Kay J D 2007 Curr. Opin. Struct. Biol. 17 3
 Google Scholar
Google Scholar
[41] Flory P J 1942 J. Chem. Phys. 10 51
 Google Scholar
Google Scholar
[42] Huggins M L 1942 J. Chem. Phys. 46 151
 Google Scholar
Google Scholar
[43] Samanta H S, Zhuravlev P I, Hinczewski M, Hori N, Chakrabarti S, Thirumalai D 2017 Soft Matter 13 3622
 Google Scholar
Google Scholar
[44] Zhou H X, Nguemaha V, Mazarakos K, Qin S B 2018 Trends Biochem. Sci. 43 499
 Google Scholar
Google Scholar
[45] Lin Y H, Forman-Kay J D, Chan H S 2016 Phys. Rev. Lett. 117 178101
 Google Scholar
Google Scholar
[46] Liu Y X, Zhang H D, Tong C H, Yang Y L 2011 Macromolecules 44 8261
 Google Scholar
Google Scholar
[47] Speck T, Menzel A M, Bialke J, Lowen H 2015 J. Chem. Phys. 142 224109
 Google Scholar
Google Scholar
[48] Burke M G, Woscholski R, Yaliraki S N 2003 Proc. Natl. Acad. Sci. U.S.A. 100 13928
 Google Scholar
Google Scholar
[49] Ruff K M, Khan S J, Pappu R V 2014 Biophys. J. 107 1226
 Google Scholar
Google Scholar
[50] Zuccato C, Valenza M, Cattaneo E 2010 Physiol. Rev. 90 905
 Google Scholar
Google Scholar
[51] Condon J E, Martin T B, Jayaraman A 2017 Soft Matter 13 2907
 Google Scholar
Google Scholar
[52] Dignon G L, Zheng W, Kim Y C, Best R B, Mittal J 2018 PLOS Comput. Biol. 14 e1005941
 Google Scholar
Google Scholar
[53] Das S, Eisen A, Lin Y H, Chan H S 2018 J. Phys. Chem. B 122 5418
 Google Scholar
Google Scholar
[54] Kapcha L H, Rossky P J 2014 J. Mol. Boil. 426 484
 Google Scholar
Google Scholar
[55] Ashbaugh H S, Hatch H W 2008 J. Am. Chem. Soc. 130 9536
 Google Scholar
Google Scholar
[56] Ghavami A, Veenhoff L M, van der Giessen E, Onck P R 2014 Biophys. J. 107 1393
 Google Scholar
Google Scholar
[57] Borgia A, Borgia M B, Bugge K, Kissling V M, Heidarsson P O, Fernandes C B, Sottini A, Soranno A, Buholzer K J, Nettels D, Kragelund B B, Best R B, Schuler B 2018 Nature 555 61
 Google Scholar
Google Scholar
[58] Wang J, Choi J M, Holehouse A S, Lee H O, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu R V, Alberti S, Hyman A A 2018 Cell 174 688
 Google Scholar
Google Scholar
[59] Song J, Gomes G N, Shi T, Gradinaru C C, Chan H S 2017 Biophys. J. 113 1012
 Google Scholar
Google Scholar
[60] Noid W G 2013 J. Chem. Phys. 139 090901
 Google Scholar
Google Scholar
[61] Voegler Smith A, Hall C K 2001 Proteins 44 344
 Google Scholar
Google Scholar
[62] Marchut A J, Hall C K 2006 Biophys. J. 90 4574
 Google Scholar
Google Scholar
[63] Marchut A J, Hall C K 2007 Proteins 66 96
 Google Scholar
Google Scholar
[64] Nguyen H D, Hall C K 2006 J. Am. Chem. Soc. 128 1890
 Google Scholar
Google Scholar
[65] Cheon M, Chang I, Hall C K 2010 Proteins 78 2950
 Google Scholar
Google Scholar
[66] Yeo J J, Huang W W, Tarakanova A, Zhang Y W, Kaplan D L, Buehler M J 2018 J. Mater. Chem. B 6 3727
 Google Scholar
Google Scholar
[67] Bereau T, Deserno M 2009 J. Chem. Phys. 130 235106
 Google Scholar
Google Scholar
[68] Rutter G O, Brown A H, Quigley D, Walsh T R, Allen M P 2015 Phys. Chem. Chem. Phys. 17 31741
 Google Scholar
Google Scholar
[69] Chen M C, Wolynes P G 2017 Proc. Natl. Acad. Sci. U.S.A. 114 4406
 Google Scholar
Google Scholar
[70] Seo M, Rauscher S, Pomes R, Tieleman D P 2012 J. Chem. Theory Comput. 8 1774
 Google Scholar
Google Scholar
[71] Miao L, Schulten K 2009 Structure 17 449
 Google Scholar
Google Scholar
[72] Monticelli L, Kandasamy S K, Periole X, Larson R G, Tieleman D P, Marrink S J 2008 J. Chem. Theory Comput. 4 819
 Google Scholar
Google Scholar
[73] Marrink S J, Risselada H J, Yefimov S, Tieleman D P, Vries A H 2007 J. Phys. Chem. B 111 7812
 Google Scholar
Google Scholar
[74] Lu L Y, Dama J F, Voth G A 2013 J. Chem. Phys. 139 121906
 Google Scholar
Google Scholar
[75] Hills R D, Lu L, Voth G A 2010 PLOS Comput. Biol. 6 e1000827
 Google Scholar
Google Scholar
[76] Ando D, Zandi R, Kim Y W, Colvin M, Rexach M, Gopinathan A J 2014 Biophys. J. 106 1997
 Google Scholar
Google Scholar
[77] Kim Y C, Hummer G 2008 J. Mol. Biol. 375 1416
 Google Scholar
Google Scholar
[78] Ryan V H, Dignon G L, Zerze G H, Chabata C V, Silva R, Conicella A E, Amaya J, Burke K A, Mittal J, Fawzi N L 2018 Mol. Cell 69 465
 Google Scholar
Google Scholar
[79] Choi J-M, Dar F, Pappu R V 2019 PLOS Comput. Biol. 15 e1007028
 Google Scholar
Google Scholar
[80] Tran H T, Mao A, Pappu R V 2008 J. Am. Chem. Soc. 130 7380
 Google Scholar
Google Scholar
[81] Holehouse A S, Garai K, Lyle N, Vitalis A, Pappu R V 2015 J. Am. Chem. Soc. 137 2984
 Google Scholar
Google Scholar
[82] Asthagiri D, Karandur D, Tomar D S, Pettitt B M 2017 J. Phys. Chem. B 121 8078
 Google Scholar
Google Scholar
[83] Karandur D, Wong K Y, Pettitt B M 2014 J. Phys. Chem. B 118 9565
 Google Scholar
Google Scholar
[84] Vitalis A, Wang X, Pappu R V 2008 J. Mol. Biol. 384 279
 Google Scholar
Google Scholar
[85] Vitalis A, Lyle N, Pappu R V 2009 Biophys. J. 97 303
 Google Scholar
Google Scholar
[86] Kang H, Vazquez F X, Zhang L, Das P, Toledo-Sherman L, Luan B, Levitt M, Zhou R 2017 J. Am. Chem. Soc. 139 8820
 Google Scholar
Google Scholar
[87] Daggett V, Kollman P A, Kuntz I D 1991 Biopolymers 31 1115
 Google Scholar
Google Scholar
计量
- 文章访问数: 23260
- PDF下载量: 530
- 被引次数: 0













 下载:
下载:
